"nuclear receptor subfamily 2, group F, member 1a"
ZFIN 
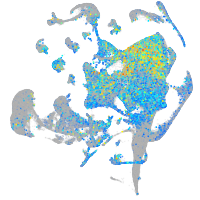
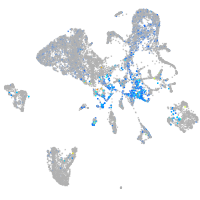
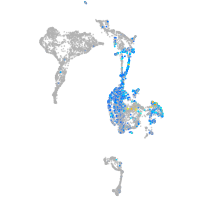
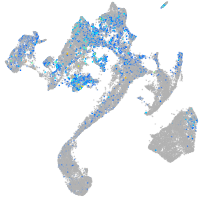
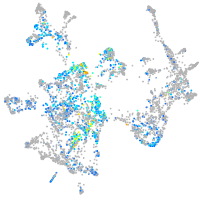
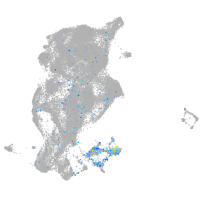
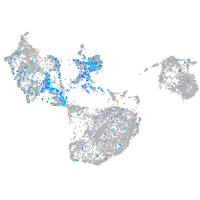
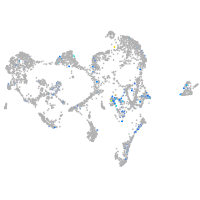
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| nova2 | 0.328 | gstp1 | -0.131 |
| elavl3 | 0.303 | gpr143 | -0.127 |
| stmn1b | 0.295 | tspan36 | -0.125 |
| nr2f2 | 0.294 | syngr1a | -0.123 |
| tuba1c | 0.285 | si:dkey-21a6.5 | -0.117 |
| tmeff1b | 0.282 | pcbd1 | -0.111 |
| tmsb | 0.272 | rab34b | -0.108 |
| ptmaa | 0.272 | LOC103910009 | -0.106 |
| gpm6aa | 0.271 | smim29 | -0.105 |
| hmgb3a | 0.268 | mitfa | -0.105 |
| zc4h2 | 0.262 | rab38 | -0.105 |
| rtn1a | 0.262 | akr1b1 | -0.103 |
| si:ch211-222l21.1 | 0.259 | qdpra | -0.103 |
| epb41a | 0.258 | actb2 | -0.101 |
| gng3 | 0.255 | vamp3 | -0.100 |
| fez1 | 0.246 | pttg1ipb | -0.098 |
| tox | 0.245 | ahnak | -0.098 |
| sox11b | 0.244 | rabl6b | -0.098 |
| hmgb1b | 0.243 | C12orf75 | -0.097 |
| tubb5 | 0.238 | prdx6 | -0.097 |
| fam168a | 0.236 | pts | -0.096 |
| marcksl1a | 0.233 | gch2 | -0.095 |
| ebf3a | 0.232 | bace2 | -0.094 |
| CU467822.1 | 0.230 | cst14a.2 | -0.093 |
| pbx1a | 0.230 | slc3a2a | -0.092 |
| sox11a | 0.229 | cdh1 | -0.092 |
| ptmab | 0.228 | si:dkey-204f11.64 | -0.092 |
| FO082781.1 | 0.227 | krt18b | -0.091 |
| marcksb | 0.226 | zgc:110339 | -0.091 |
| rnasekb | 0.224 | tspan10 | -0.090 |
| chd4a | 0.224 | slc45a2 | -0.089 |
| si:ch73-1a9.3 | 0.223 | zgc:110239 | -0.088 |
| pbx3b | 0.221 | pfn1 | -0.088 |
| elavl4 | 0.220 | slc38a5b | -0.087 |
| sncb | 0.217 | bloc1s6 | -0.087 |