"nuclear receptor subfamily 0, group B, member 2a"
ZFIN 
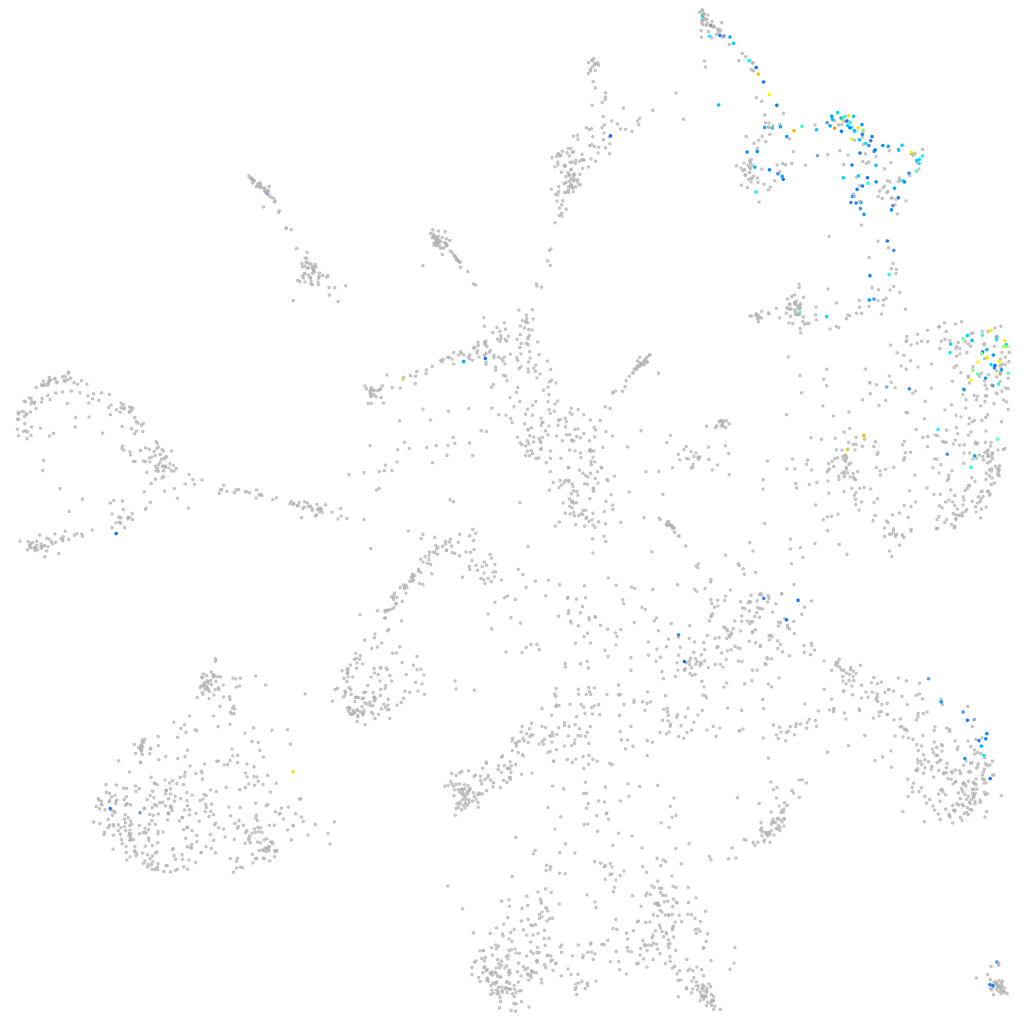
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| alpi.1 | 0.386 | BX088707.3 | -0.185 |
| sfrp5 | 0.343 | tpm1 | -0.172 |
| mmel1 | 0.337 | myl9a | -0.163 |
| synpo2lb | 0.325 | acta2 | -0.155 |
| cxadr | 0.318 | ptmaa | -0.155 |
| podxl | 0.313 | tagln | -0.144 |
| lmod2a | 0.312 | myh11a | -0.130 |
| fabp11a | 0.291 | gapdhs | -0.128 |
| gata5 | 0.289 | myl6 | -0.126 |
| COLEC10 | 0.287 | mylkb | -0.125 |
| crb2a | 0.287 | foxf1 | -0.114 |
| tnni1b | 0.285 | tpm2 | -0.112 |
| lbx2 | 0.279 | ntn5 | -0.112 |
| slc29a1b | 0.278 | mdkb | -0.111 |
| tmem88b | 0.271 | lmod1b | -0.110 |
| angptl6 | 0.266 | si:ch211-62a1.3 | -0.109 |
| krt94 | 0.266 | si:dkeyp-66d1.7 | -0.108 |
| jam2b | 0.261 | nkx2.3 | -0.108 |
| krt8 | 0.261 | pbx3b | -0.107 |
| aqp1a.1 | 0.253 | ckbb | -0.107 |
| si:ch211-214p16.2 | 0.251 | XLOC-041870 | -0.103 |
| krt18a.1 | 0.251 | inka1a | -0.103 |
| bnc2 | 0.250 | zgc:153704 | -0.102 |
| alx4a | 0.249 | foxp4 | -0.102 |
| gstm.3 | 0.245 | XLOC-025423 | -0.100 |
| ncs1a | 0.240 | csrp1b | -0.100 |
| tbx5a | 0.237 | ppiab | -0.099 |
| gata6 | 0.230 | col5a2a | -0.096 |
| hsd3b7 | 0.228 | col1a2 | -0.095 |
| bcam | 0.226 | cald1b | -0.095 |
| jcada | 0.222 | ak1 | -0.094 |
| thy1 | 0.220 | desmb | -0.093 |
| tjp2b | 0.219 | fhl2a | -0.091 |
| arl4aa | 0.217 | myocd | -0.090 |
| clec19a | 0.216 | pmp22a | -0.089 |