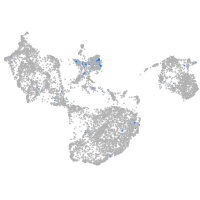
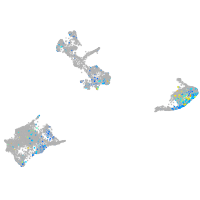
neuronal pentraxin 1 like
ZFIN 
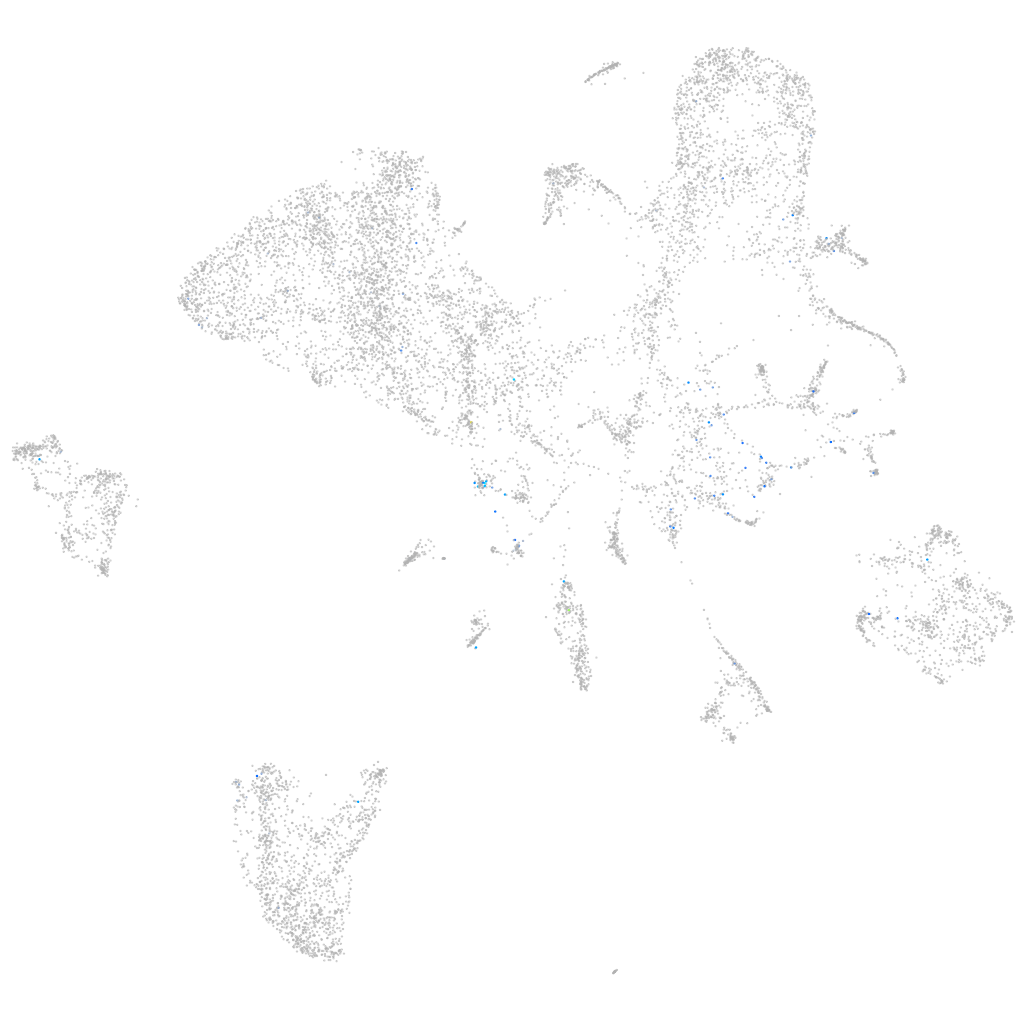
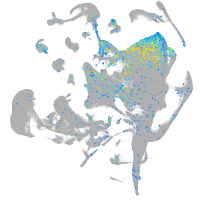
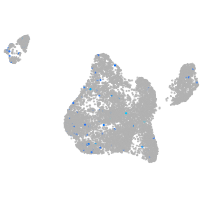
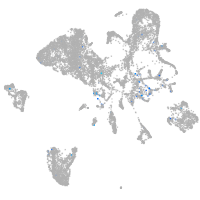
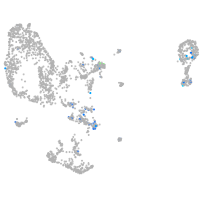
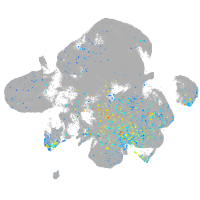
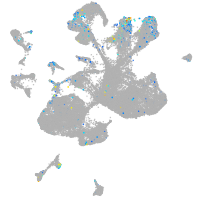
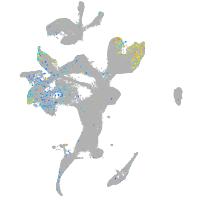
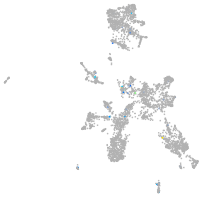
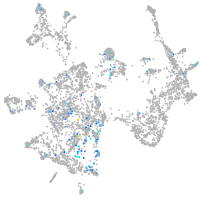
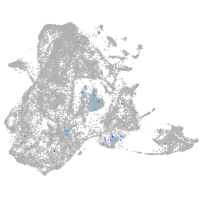
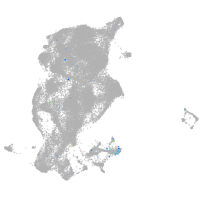
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| XLOC-006820 | 0.293 | aldob | -0.050 |
| si:ch73-233f7.6 | 0.291 | sod2 | -0.049 |
| BX936415.1 | 0.290 | scp2a | -0.048 |
| CU861477.1 | 0.279 | ahcy | -0.047 |
| trpc6b | 0.247 | gapdh | -0.046 |
| gpr88 | 0.235 | sod1 | -0.046 |
| XLOC-014755 | 0.204 | gstt1a | -0.045 |
| zgc:153118 | 0.202 | gstp1 | -0.044 |
| myadml2 | 0.199 | aldh6a1 | -0.044 |
| CR847968.1 | 0.198 | cx32.3 | -0.044 |
| XLOC-036573 | 0.196 | sdr16c5b | -0.043 |
| XLOC-013339 | 0.190 | mgst1.2 | -0.043 |
| frmpd3 | 0.189 | ckba | -0.043 |
| pcdh1gc6 | 0.184 | glud1b | -0.043 |
| LOC110440219 | 0.184 | eno3 | -0.043 |
| rtn1b | 0.183 | eef1da | -0.042 |
| LOC103911776.1 | 0.182 | gcshb | -0.042 |
| desmb | 0.180 | chchd10 | -0.042 |
| cadm3 | 0.179 | fdx1 | -0.042 |
| LOC101885553 | 0.177 | mat1a | -0.042 |
| ank3a | 0.177 | pgk1 | -0.042 |
| lingo2a | 0.171 | nipsnap3a | -0.042 |
| cacng2a | 0.170 | ugt1a7 | -0.041 |
| atn1 | 0.170 | pnp4b | -0.040 |
| kcnq3 | 0.170 | mt-nd3 | -0.040 |
| dnajb5 | 0.168 | gamt | -0.040 |
| si:ch73-379j16.2 | 0.167 | eci2 | -0.040 |
| XLOC-015481 | 0.165 | slco1d1 | -0.040 |
| zgc:153426 | 0.162 | pgm1 | -0.040 |
| cacna1g | 0.162 | nupr1b | -0.039 |
| celf4 | 0.162 | agxtb | -0.039 |
| gabrd | 0.161 | slc25a20 | -0.039 |
| mturn | 0.159 | pklr | -0.039 |
| gdap1l1 | 0.159 | rdh1 | -0.039 |
| LOC110438360 | 0.159 | g6pca.2 | -0.039 |