"Niemann-Pick disease, type C2"
ZFIN 
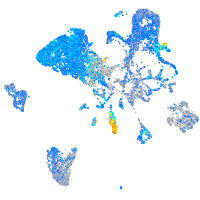
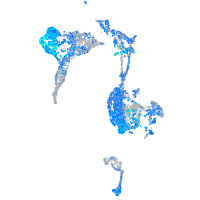
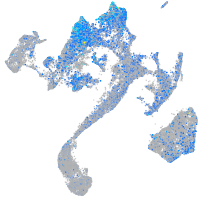
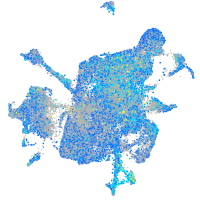
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| tspan36 | 0.290 | ptmab | -0.167 |
| slc45a2 | 0.281 | si:ch73-1a9.3 | -0.133 |
| dct | 0.278 | hmgn6 | -0.103 |
| sparc | 0.275 | epb41a | -0.092 |
| gpr143 | 0.275 | nova2 | -0.091 |
| pmela | 0.273 | ptmaa | -0.086 |
| cd63 | 0.271 | gpm6aa | -0.084 |
| tyrp1b | 0.270 | tuba1c | -0.079 |
| pmelb | 0.253 | hmgb1a | -0.073 |
| gstp1 | 0.250 | snap25b | -0.073 |
| tyrp1a | 0.249 | neurod4 | -0.072 |
| tspan10 | 0.248 | hmgb3a | -0.071 |
| bace2 | 0.248 | si:ch211-137a8.4 | -0.071 |
| rab38 | 0.247 | calb2b | -0.071 |
| pttg1ipb | 0.244 | anp32e | -0.070 |
| col4a5 | 0.242 | ndrg4 | -0.070 |
| agtrap | 0.242 | vsx1 | -0.069 |
| rnaseka | 0.240 | marcksl1b | -0.067 |
| col4a6 | 0.238 | syt5b | -0.067 |
| slc24a5 | 0.236 | gnb3a | -0.065 |
| zgc:110591 | 0.234 | tuba1a | -0.065 |
| tyr | 0.230 | crx | -0.064 |
| mitfa | 0.229 | mpp6b | -0.064 |
| oca2 | 0.228 | si:dkey-276j7.1 | -0.063 |
| cx43 | 0.228 | golga7ba | -0.063 |
| qdpra | 0.228 | zc4h2 | -0.063 |
| fstl1b | 0.227 | gng13b | -0.062 |
| pcbd1 | 0.227 | nrn1lb | -0.061 |
| rgrb | 0.227 | cadm3 | -0.061 |
| cracr2ab | 0.227 | pcbp3 | -0.060 |
| fabp11b | 0.220 | atp1b2a | -0.060 |
| rab32a | 0.219 | vamp1 | -0.059 |
| trpm1b | 0.218 | atp1a3a | -0.059 |
| slc3a2a | 0.218 | gnao1b | -0.058 |
| tmem98 | 0.213 | scrt2 | -0.058 |