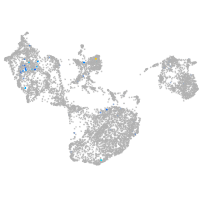
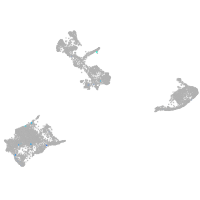
neuropeptide B
ZFIN 
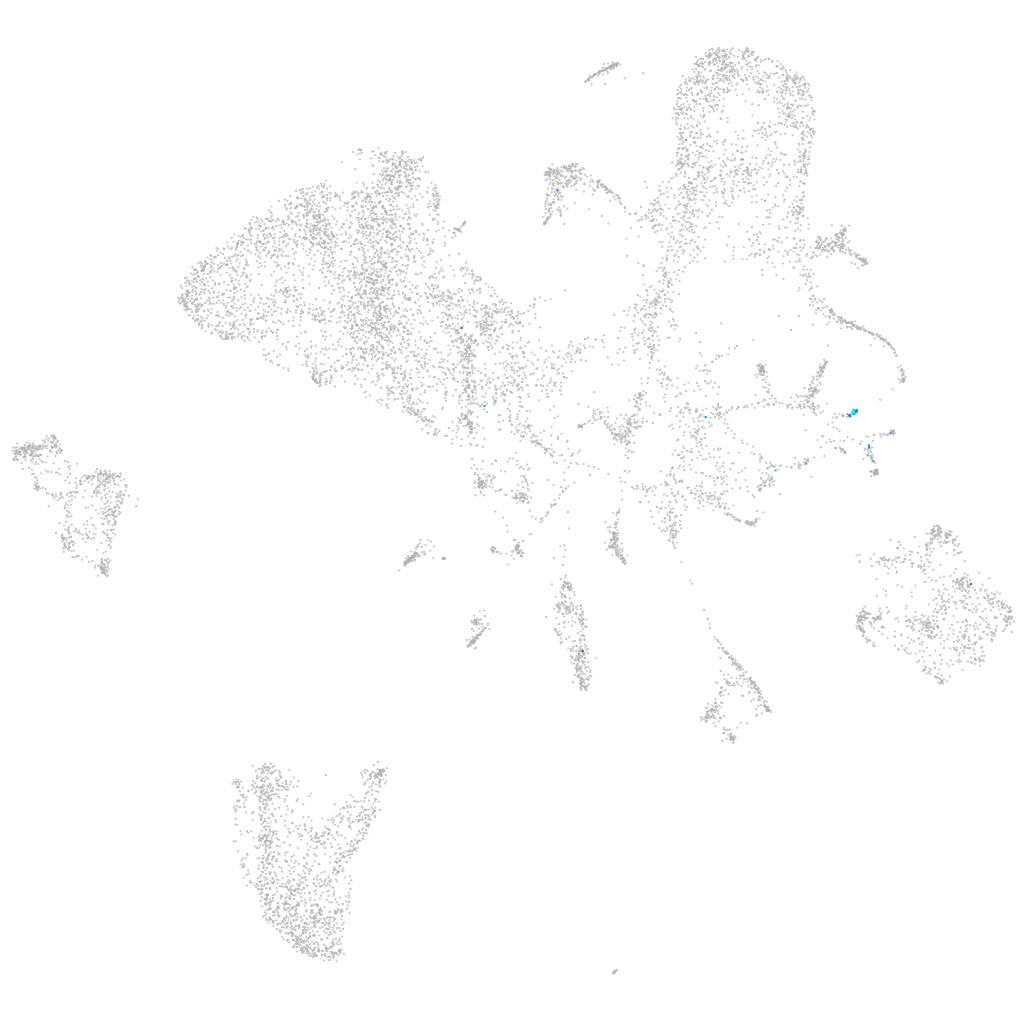
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| pnoca | 0.718 | gapdh | -0.063 |
| gcgb | 0.636 | hdlbpa | -0.056 |
| gcga | 0.566 | ahcy | -0.055 |
| ak5l | 0.487 | gstp1 | -0.054 |
| dkk3b | 0.445 | aldh6a1 | -0.053 |
| cntnap2a | 0.348 | gatm | -0.050 |
| slc30a2 | 0.341 | atp1a1a.1 | -0.050 |
| plppr5b | 0.335 | aldh9a1a.1 | -0.050 |
| adora2ab | 0.328 | cox6b1 | -0.050 |
| rims2a | 0.327 | bhmt | -0.049 |
| ptn | 0.321 | ctsla | -0.049 |
| LOC571123 | 0.311 | mt-nd4 | -0.049 |
| elfn1a | 0.282 | eno3 | -0.048 |
| rprmb | 0.271 | ssr3 | -0.048 |
| rims4 | 0.266 | gstr | -0.046 |
| chrnb3b | 0.265 | mat1a | -0.046 |
| lgi2a | 0.254 | glud1b | -0.046 |
| dgkh | 0.250 | aqp12 | -0.046 |
| gpsm1a | 0.244 | ckba | -0.046 |
| CR774179.5 | 0.239 | sdr16c5b | -0.045 |
| zgc:194312 | 0.234 | tuba8l2 | -0.045 |
| scgn | 0.231 | gamt | -0.045 |
| disp2 | 0.225 | nupr1b | -0.045 |
| si:ch73-160i9.2 | 0.222 | cebpb | -0.044 |
| adgrb2 | 0.216 | apoa1b | -0.044 |
| cfap58 | 0.212 | gpx1a | -0.043 |
| gpr142 | 0.209 | cx32.3 | -0.043 |
| gsg1l2b | 0.208 | jun | -0.043 |
| adm2a | 0.202 | lgals2b | -0.043 |
| SHISAL2A | 0.202 | apoa4b.1 | -0.042 |
| kansl1l | 0.198 | prdx2 | -0.042 |
| pax6b | 0.198 | rps20 | -0.042 |
| gfra3 | 0.196 | mt-atp6 | -0.042 |
| rgs5a | 0.194 | agxta | -0.042 |
| fap | 0.193 | egr1 | -0.041 |