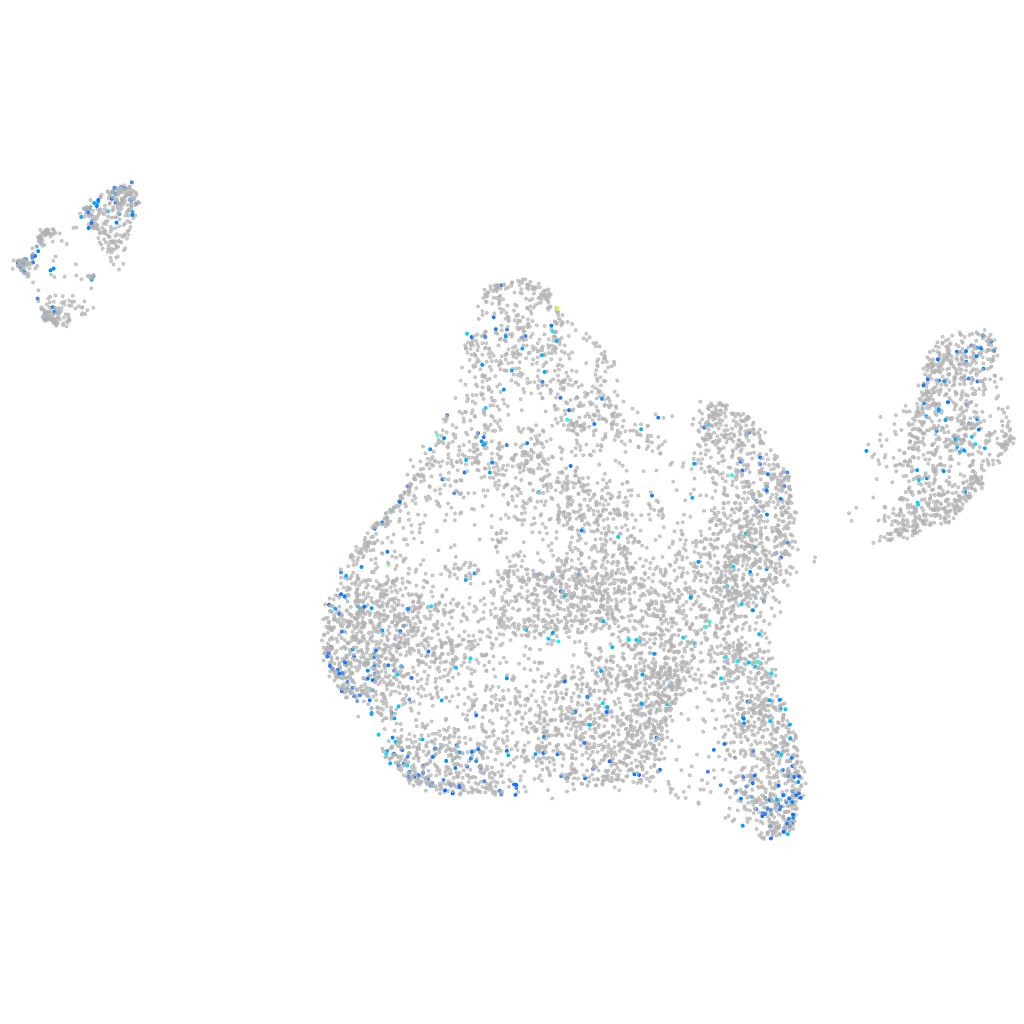
NADPH oxidase activator 1
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm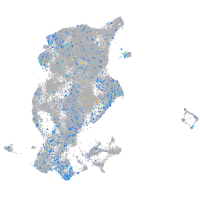 |
fin |
ionocytes / mucous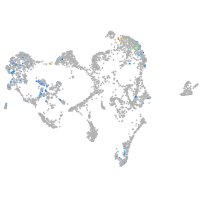 |
pigment cells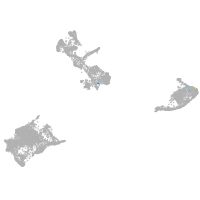 |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| CR550302.1 | 0.101 | hmgn2 | -0.062 |
| CR788311.1 | 0.087 | hmgn6 | -0.056 |
| hcn1 | 0.081 | NC-002333.4 | -0.055 |
| si:dkey-174i8.1 | 0.077 | hmgb2a | -0.051 |
| cdk5r1a | 0.076 | hspb1 | -0.051 |
| BFSP1 | 0.075 | crabp2b | -0.047 |
| capn5b | 0.075 | hnrnpa0b | -0.045 |
| prkcg | 0.075 | si:ch73-1a9.3 | -0.044 |
| dgcr8 | 0.073 | si:ch211-222l21.1 | -0.040 |
| zp3 | 0.070 | sox2 | -0.040 |
| LO018598.1 | 0.070 | cox7b | -0.039 |
| CU681855.1 | 0.070 | ptmab | -0.037 |
| XLOC-036507 | 0.070 | ccng2 | -0.036 |
| gpr37b | 0.070 | marcksl1b | -0.036 |
| BEND4 | 0.069 | ube2e2 | -0.035 |
| pcdh1g2 | 0.069 | apoeb | -0.034 |
| swap70a | 0.069 | cdx4 | -0.033 |
| LOC103909472 | 0.069 | slc25a5 | -0.033 |
| fam20ca | 0.069 | ptgdsb.1 | -0.033 |
| h1m | 0.069 | cdkn1cb | -0.032 |
| dram2a | 0.068 | add3b | -0.032 |
| mov10b.1 | 0.068 | BX927258.1 | -0.031 |
| irge4 | 0.068 | xk | -0.031 |
| tppp3 | 0.067 | rplp0 | -0.031 |
| ehmt1a | 0.067 | ptger4b | -0.030 |
| vtg1 | 0.067 | lhx5 | -0.030 |
| XLOC-043679 | 0.067 | urb1 | -0.029 |
| pimr83 | 0.067 | birc5a | -0.029 |
| gyg1a | 0.067 | atp1b2a | -0.029 |
| mycla | 0.067 | hnrnpa0a | -0.029 |
| col4a6 | 0.067 | LOC108190024 | -0.029 |
| si:ch211-214k5.3 | 0.066 | atp1b3a | -0.028 |
| dapp1 | 0.065 | lmo4a | -0.028 |
| myct1a | 0.065 | rps13 | -0.028 |
| tnni1c | 0.065 | apoc1 | -0.027 |