NOVA alternative splicing regulator 2
ZFIN 
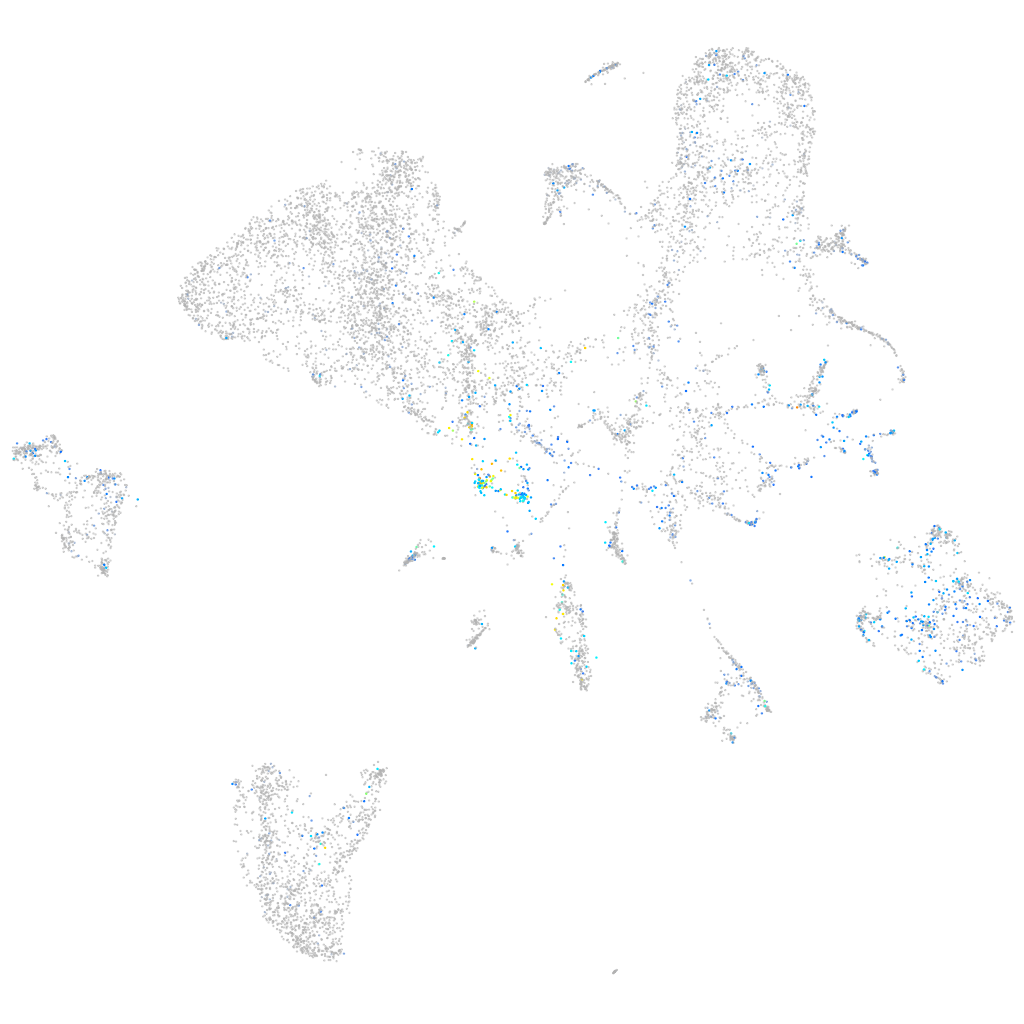
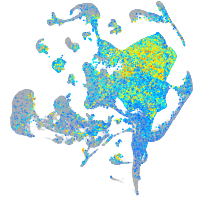
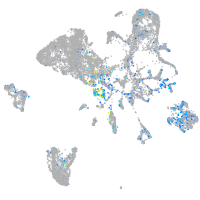
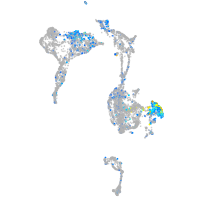
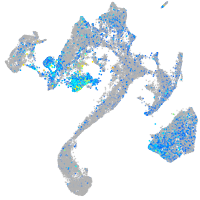
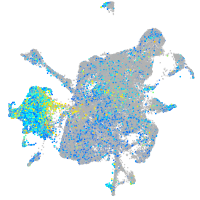
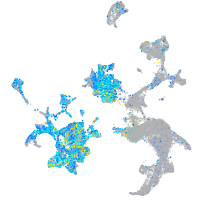
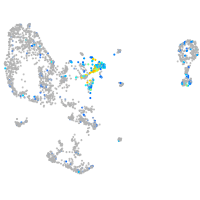
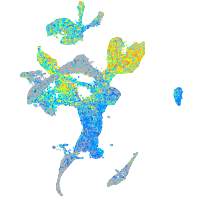
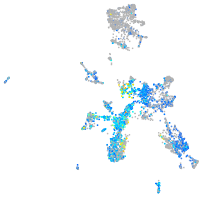
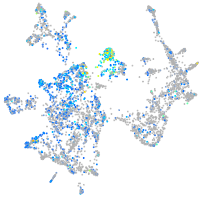
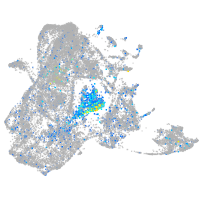
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| elavl3 | 0.423 | gapdh | -0.183 |
| tuba1c | 0.389 | eno3 | -0.181 |
| gpm6aa | 0.355 | glud1b | -0.171 |
| tuba1a | 0.325 | aldob | -0.170 |
| stmn1b | 0.322 | sod1 | -0.170 |
| cspg5a | 0.322 | ahcy | -0.169 |
| mdkb | 0.317 | eef1da | -0.167 |
| rtn1b | 0.304 | aldh6a1 | -0.166 |
| si:dkey-276j7.1 | 0.303 | gamt | -0.165 |
| tmeff1b | 0.288 | fbp1b | -0.164 |
| tubb5 | 0.286 | nupr1b | -0.161 |
| hmgb3a | 0.280 | ckba | -0.159 |
| cadm3 | 0.270 | cx32.3 | -0.158 |
| celf2 | 0.268 | mat1a | -0.156 |
| myt1la | 0.266 | scp2a | -0.155 |
| myt1a | 0.265 | gstr | -0.152 |
| ncam1a | 0.264 | dap | -0.150 |
| gng3 | 0.262 | pklr | -0.150 |
| marcksl1b | 0.262 | gstt1a | -0.149 |
| hmgb1b | 0.259 | pnp4b | -0.149 |
| dpysl3 | 0.257 | cebpd | -0.148 |
| si:ch211-137a8.4 | 0.255 | gatm | -0.148 |
| FO082781.1 | 0.253 | suclg1 | -0.145 |
| ebf3a | 0.252 | aldh7a1 | -0.144 |
| mab21l2 | 0.252 | abat | -0.143 |
| zc4h2 | 0.252 | atp5mc3b | -0.143 |
| CU467822.1 | 0.251 | hdlbpa | -0.142 |
| epb41a | 0.250 | rdh1 | -0.142 |
| myt1b | 0.248 | agxta | -0.142 |
| XLOC-003690 | 0.247 | akt2l | -0.141 |
| dpysl2b | 0.246 | agxtb | -0.141 |
| XLOC-003689 | 0.246 | acadm | -0.140 |
| elavl4 | 0.246 | gcshb | -0.139 |
| CU634008.1 | 0.246 | gpx4a | -0.139 |
| scrt2 | 0.245 | eci2 | -0.138 |