nitric oxide synthase trafficking
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm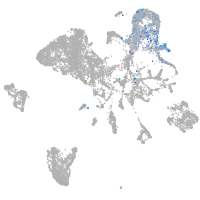 |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural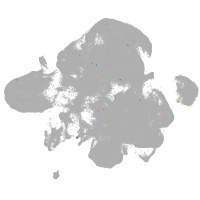 |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin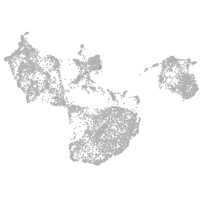 |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| pdzk1 | 0.355 | fabp3 | -0.040 |
| lrrc66 | 0.352 | marcksl1a | -0.038 |
| eps8l3b | 0.344 | atp6v0cb | -0.033 |
| eps8l3a | 0.326 | tuba1c | -0.033 |
| amn | 0.323 | rtn1a | -0.032 |
| si:dkey-194e6.1 | 0.323 | gpm6aa | -0.031 |
| cdh17 | 0.322 | gpm6ab | -0.031 |
| si:ch73-185c24.2 | 0.313 | nova2 | -0.031 |
| ezra | 0.296 | actc1b | -0.029 |
| bin2a | 0.294 | celf2 | -0.029 |
| CU682777.2 | 0.292 | mdkb | -0.029 |
| muc13b | 0.292 | rnasekb | -0.029 |
| pls1 | 0.291 | tubb5 | -0.029 |
| si:dkey-103j14.5 | 0.291 | gapdhs | -0.028 |
| IZUMO1R | 0.290 | jpt1b | -0.028 |
| atp1a1a.4 | 0.289 | CABZ01080702.1 | -0.027 |
| ggt1b | 0.288 | fam168a | -0.027 |
| slc6a19a.1 | 0.286 | mdka | -0.027 |
| sdsl | 0.272 | hmgb1b | -0.026 |
| si:ch211-274f20.2 | 0.265 | tmeff1b | -0.026 |
| slc5a9 | 0.265 | atp1b2a | -0.025 |
| zgc:101040 | 0.264 | cadm3 | -0.025 |
| enpep | 0.263 | cspg5a | -0.025 |
| si:ch211-117n7.7 | 0.263 | rtn1b | -0.025 |
| epdl2 | 0.262 | atpv0e2 | -0.024 |
| si:dkey-30c15.13 | 0.260 | elavl3 | -0.024 |
| mbl2 | 0.257 | hmgb3a | -0.024 |
| cox6a2 | 0.255 | sncb | -0.024 |
| slc6a19a.2 | 0.255 | tnrc6c1 | -0.024 |
| smim1 | 0.250 | tuba1a | -0.024 |
| st3gal7 | 0.250 | atp1a3a | -0.023 |
| cldn15a | 0.250 | cd99l2 | -0.023 |
| slc26a6 | 0.249 | hnrnpa0a | -0.023 |
| si:ch211-139a5.9 | 0.249 | stmn1b | -0.023 |
| hnf4g | 0.248 | sypb | -0.023 |




