"non-POU domain containing, octamer-binding"
ZFIN 
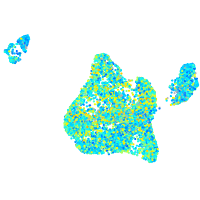
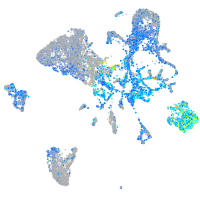
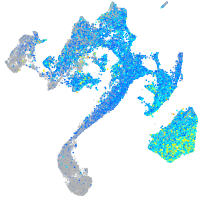
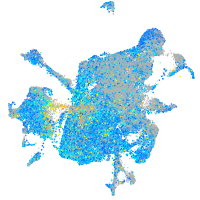
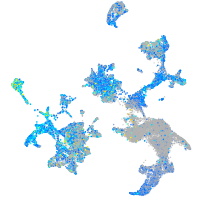
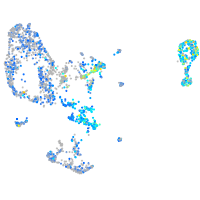
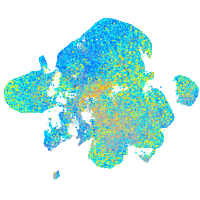
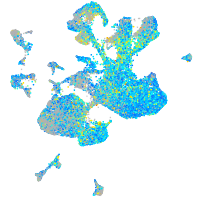
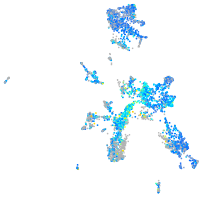
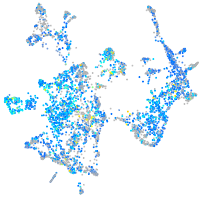
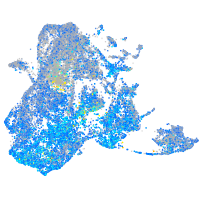
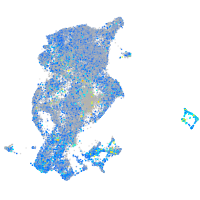
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hnrnpabb | 0.664 | actc1b | -0.576 |
| hmga1a | 0.663 | ckma | -0.530 |
| hnrnpaba | 0.657 | ckmb | -0.530 |
| khdrbs1a | 0.656 | ak1 | -0.529 |
| hmgb2a | 0.654 | atp2a1 | -0.524 |
| hnrnpa0b | 0.644 | ttn.2 | -0.518 |
| si:ch211-222l21.1 | 0.634 | aldoab | -0.515 |
| hmgb2b | 0.634 | tnnc2 | -0.509 |
| seta | 0.625 | neb | -0.504 |
| cirbpa | 0.625 | ttn.1 | -0.495 |
| cbx3a | 0.623 | tmem38a | -0.491 |
| anp32b | 0.617 | ldb3b | -0.484 |
| h2afvb | 0.615 | acta1b | -0.484 |
| nucks1a | 0.615 | mylpfa | -0.481 |
| si:ch73-281n10.2 | 0.615 | pabpc4 | -0.480 |
| syncrip | 0.615 | si:ch73-367p23.2 | -0.476 |
| si:ch73-1a9.3 | 0.614 | nme2b.2 | -0.475 |
| setb | 0.606 | actn3b | -0.475 |
| ptmab | 0.601 | actn3a | -0.475 |
| cx43.4 | 0.600 | tpma | -0.474 |
| si:ch211-288g17.3 | 0.597 | eno3 | -0.471 |
| hdac1 | 0.596 | gapdh | -0.470 |
| anp32a | 0.595 | ldb3a | -0.469 |
| rbm8a | 0.592 | myom1a | -0.465 |
| srsf3b | 0.590 | CABZ01078594.1 | -0.461 |
| marcksb | 0.587 | mybphb | -0.459 |
| cirbpb | 0.587 | mylz3 | -0.459 |
| ncl | 0.583 | si:ch211-266g18.10 | -0.459 |
| top1l | 0.583 | srl | -0.459 |
| hnrnpa1b | 0.582 | smyd1a | -0.458 |
| snrpb | 0.582 | tnnt3a | -0.458 |
| nop58 | 0.582 | ank1a | -0.455 |
| ilf3b | 0.575 | cav3 | -0.455 |
| h3f3d | 0.573 | eno1a | -0.454 |
| hmgn7 | 0.570 | tmod4 | -0.453 |