NME/NM23 family member 5
ZFIN 
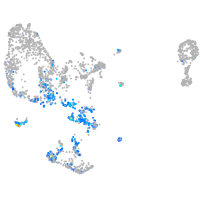
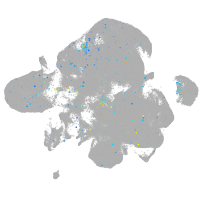
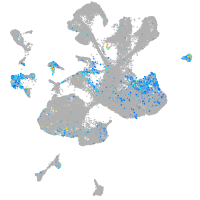
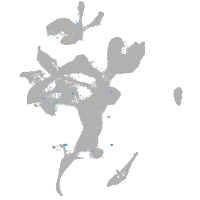
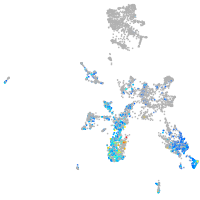
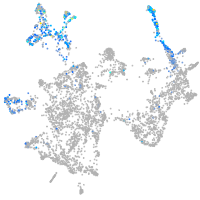
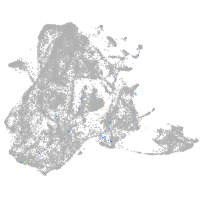
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ccdc65 | 0.220 | tnnc2 | -0.027 |
| znf1085 | 0.205 | rplp2 | -0.027 |
| cfap157 | 0.200 | ak1 | -0.026 |
| smkr1 | 0.184 | myom1a | -0.026 |
| CABZ01043826.1 | 0.180 | cfl2 | -0.026 |
| si:dkeyp-110a12.4 | 0.171 | eno1a | -0.025 |
| ANO2 (1 of many) | 0.170 | gapdh | -0.025 |
| CSDC2 | 0.169 | ckmb | -0.025 |
| drc3 | 0.158 | ldb3a | -0.025 |
| ak7a | 0.157 | si:ch73-367p23.2 | -0.025 |
| daw1 | 0.149 | ldb3b | -0.025 |
| si:dkey-27p23.3 | 0.141 | actn3a | -0.024 |
| dydc2 | 0.138 | eef2l2 | -0.024 |
| XLOC-025509 | 0.133 | si:ch211-266g18.10 | -0.024 |
| spag8 | 0.129 | acta1b | -0.024 |
| plpp4 | 0.128 | ckma | -0.024 |
| morn3 | 0.125 | pabpc4 | -0.024 |
| zswim2 | 0.122 | ttn.1 | -0.024 |
| si:ch211-93e11.8 | 0.120 | XLOC-025819 | -0.024 |
| lrrc7 | 0.119 | rtn2a | -0.024 |
| ccdc173 | 0.119 | atp2a1 | -0.024 |
| vwa5b1 | 0.119 | tpma | -0.023 |
| nitr12 | 0.117 | pgam2 | -0.023 |
| CABZ01085140.1 | 0.117 | casq1b | -0.023 |
| lrrc74b | 0.117 | mybphb | -0.023 |
| pcdh15a | 0.116 | pkmb | -0.023 |
| acot21 | 0.115 | aldoab | -0.023 |
| si:dkey-163f12.6 | 0.114 | actc1b | -0.023 |
| bcan | 0.111 | mylpfa | -0.023 |
| dnali1 | 0.109 | mylz3 | -0.023 |
| si:ch211-163l21.7 | 0.109 | tmod4 | -0.023 |
| LOC108183289 | 0.109 | atp1a2a | -0.023 |
| spata17 | 0.108 | prr33 | -0.023 |
| ccdc151 | 0.107 | dhrs7cb | -0.023 |
| dnaaf3 | 0.106 | myom2a | -0.023 |