"NME/NM23 nucleoside diphosphate kinase 2b, tandem duplicate 2"
ZFIN 
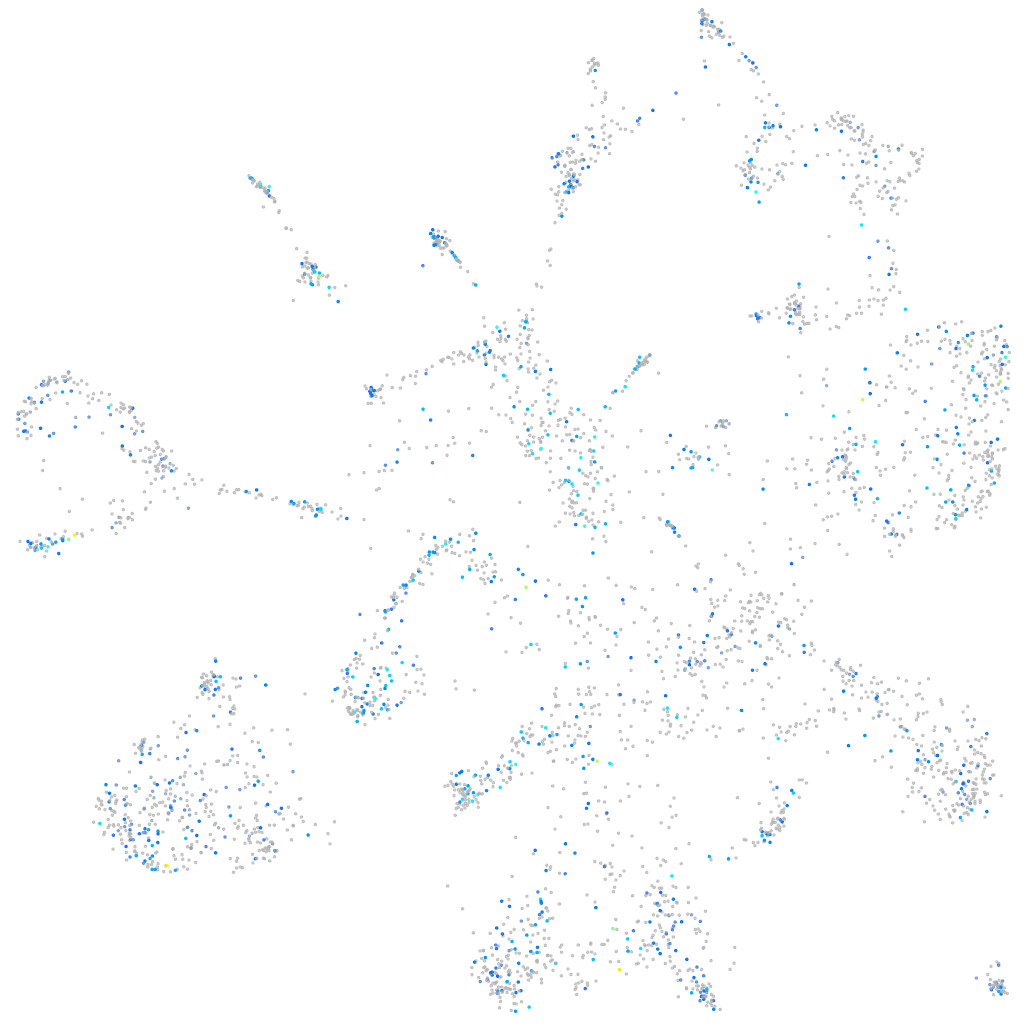
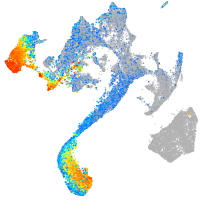
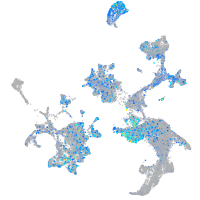
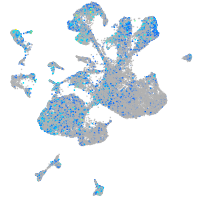
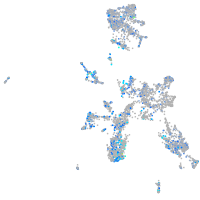
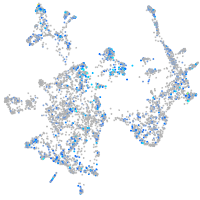
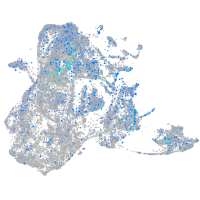
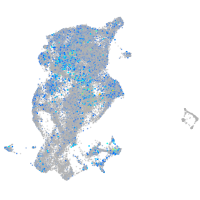
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| pvalb2 | 0.276 | rplp2l | -0.154 |
| actc1b | 0.244 | rps12 | -0.150 |
| pvalb1 | 0.241 | rps8a | -0.137 |
| mylz3 | 0.238 | rpl36 | -0.136 |
| mylpfa | 0.205 | rps16 | -0.127 |
| apoa2 | 0.202 | rpl18a | -0.125 |
| tnni2a.4 | 0.192 | cfl1 | -0.125 |
| tnnt3b | 0.187 | si:ch211-222l21.1 | -0.125 |
| apoa1b | 0.170 | rps24 | -0.124 |
| pvalb4 | 0.159 | uba52 | -0.121 |
| myhz1.1 | 0.151 | rpl38 | -0.119 |
| tpma | 0.146 | zgc:114188 | -0.119 |
| ckma | 0.142 | rpl23 | -0.117 |
| zgc:136461 | 0.142 | rpl35a | -0.117 |
| myl1 | 0.140 | seta | -0.113 |
| mylpfb | 0.137 | h2afvb | -0.113 |
| ctrb1 | 0.132 | rpl31 | -0.113 |
| CELA1 (1 of many) | 0.131 | rack1 | -0.110 |
| krt4 | 0.131 | rps10 | -0.109 |
| icn | 0.130 | rps11 | -0.109 |
| myhz2 | 0.126 | rpl23a | -0.107 |
| prss59.2 | 0.125 | sub1a | -0.107 |
| myl13 | 0.124 | pabpc1a | -0.106 |
| myl10 | 0.122 | rps18 | -0.106 |
| si:dkey-33c14.3 | 0.121 | rps5 | -0.105 |
| ttn.2 | 0.120 | rpl36a | -0.104 |
| gapdh | 0.118 | top1l | -0.103 |
| apoa4b.1 | 0.117 | ptmab | -0.103 |
| cyt1 | 0.116 | rps6 | -0.102 |
| ckmb | 0.116 | tma7 | -0.102 |
| prss1 | 0.115 | rpl17 | -0.102 |
| rbp4 | 0.108 | snrpb | -0.101 |
| krt17 | 0.107 | rps21 | -0.101 |
| fabp10a | 0.105 | rps3 | -0.100 |
| prss59.1 | 0.105 | rpl15 | -0.100 |