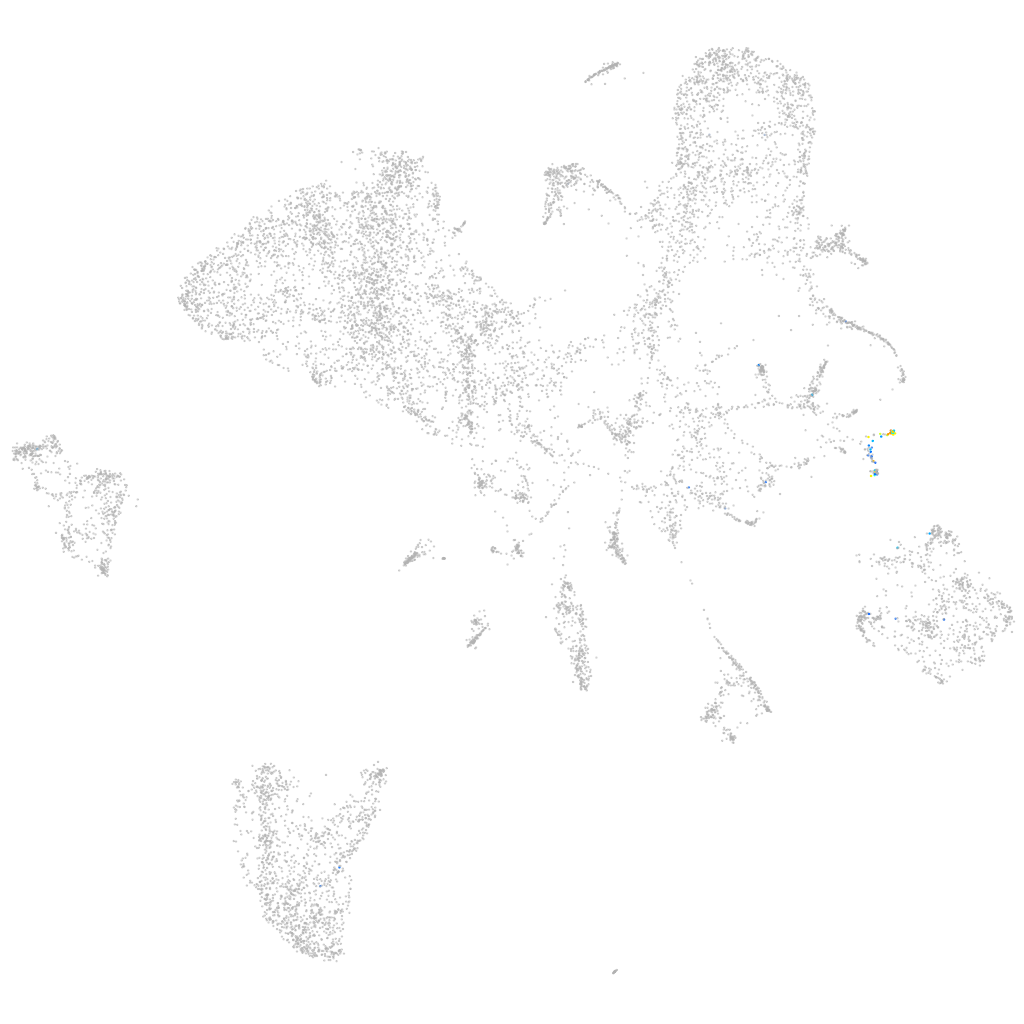
neuromedin Ba
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| zgc:195023 | 0.716 | rps20 | -0.090 |
| sst2 | 0.716 | ahcy | -0.088 |
| CU019662.1 | 0.543 | aldob | -0.084 |
| sst1.2 | 0.512 | gapdh | -0.079 |
| kctd12.2 | 0.487 | eno3 | -0.071 |
| adgb | 0.451 | gstp1 | -0.068 |
| pdyn | 0.448 | aldh9a1a.1 | -0.067 |
| gldn | 0.424 | gamt | -0.066 |
| gpr4 | 0.415 | prdx2 | -0.065 |
| scg2b | 0.389 | gstr | -0.064 |
| casr | 0.376 | glud1b | -0.062 |
| nrcama | 0.364 | adh5 | -0.061 |
| stc1l | 0.363 | fbp1b | -0.060 |
| kcnt1 | 0.343 | gstt1a | -0.060 |
| map3k15 | 0.330 | ndufa4l | -0.060 |
| slc43a2a | 0.318 | gatm | -0.059 |
| napbb | 0.315 | fabp3 | -0.059 |
| hhex | 0.299 | gsta.1 | -0.059 |
| si:ch211-242e8.1 | 0.295 | aldh6a1 | -0.058 |
| kcnk3b | 0.294 | bhmt | -0.058 |
| lamc2 | 0.290 | nme2b.1 | -0.058 |
| LOC571123 | 0.286 | nupr1b | -0.057 |
| kl | 0.284 | apoa1b | -0.057 |
| scg2a | 0.278 | prdx6 | -0.057 |
| isl1 | 0.271 | eef1da | -0.057 |
| scgn | 0.267 | acadm | -0.057 |
| olfm2a | 0.264 | apoa4b.1 | -0.057 |
| scg3 | 0.264 | hdlbpa | -0.056 |
| gpc1a | 0.263 | apoc2 | -0.056 |
| spegb | 0.261 | ckba | -0.055 |
| tent5d | 0.260 | gpx4a | -0.055 |
| pax6b | 0.260 | rmdn1 | -0.055 |
| prlra | 0.258 | fabp2 | -0.054 |
| fam49bb | 0.255 | mat1a | -0.054 |
| ACVR1C | 0.250 | afp4 | -0.054 |