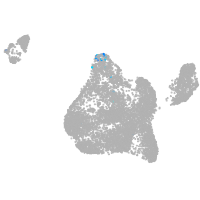
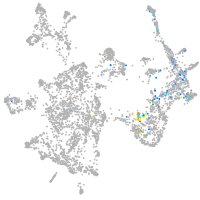
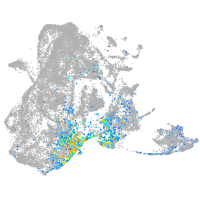
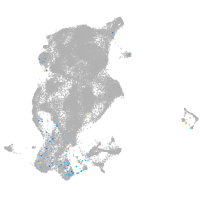
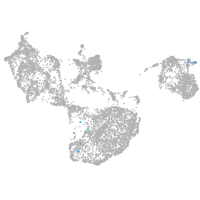
NK2 transcription factor related 7
ZFIN 
Other cell groups


Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cxcl12b | 0.400 | rpl37 | -0.230 |
| cyp26c1 | 0.387 | rps10 | -0.228 |
| mcama | 0.327 | zgc:114188 | -0.214 |
| gbx2 | 0.318 | rps17 | -0.194 |
| irx7 | 0.294 | nme2b.1 | -0.161 |
| pcdh10b | 0.283 | ahcy | -0.142 |
| rarga | 0.274 | eno3 | -0.133 |
| si:ch211-152c2.3 | 0.266 | atp5fa1 | -0.128 |
| smoc2 | 0.265 | atp5if1b | -0.127 |
| fscn2b | 0.261 | sod1 | -0.127 |
| XLOC-028299 | 0.260 | prdx2 | -0.125 |
| tbx1 | 0.246 | gapdh | -0.125 |
| sox3 | 0.244 | eef1da | -0.124 |
| akap12b | 0.239 | zgc:158463 | -0.124 |
| apela | 0.238 | zgc:92744 | -0.122 |
| lima1a | 0.238 | zgc:56493 | -0.118 |
| klf17 | 0.237 | tpi1b | -0.118 |
| marcksb | 0.232 | suclg1 | -0.117 |
| pltp | 0.225 | atp5mc1 | -0.115 |
| cox4i2 | 0.223 | atp5l | -0.114 |
| hspb1 | 0.223 | atp5f1b | -0.114 |
| NC-002333.4 | 0.218 | gamt | -0.112 |
| id1 | 0.216 | mt-nd1 | -0.112 |
| fzd7a | 0.214 | COX7A2 (1 of many) | -0.110 |
| asph | 0.212 | dap | -0.108 |
| COX7A2 | 0.210 | gstp1 | -0.108 |
| ctnnb1 | 0.209 | mdh1aa | -0.107 |
| marcksl1b | 0.205 | gpx4a | -0.105 |
| hnrnpa1a | 0.205 | mt-atp6 | -0.104 |
| ppig | 0.202 | pklr | -0.104 |
| cdkn1ca | 0.195 | lgals2b | -0.103 |
| nucks1a | 0.194 | ndufa3 | -0.103 |
| anp32a | 0.194 | cox6b1 | -0.102 |
| acin1a | 0.193 | glud1b | -0.101 |
| vox | 0.191 | gatm | -0.101 |