"nitrilase family, member 2"
ZFIN 
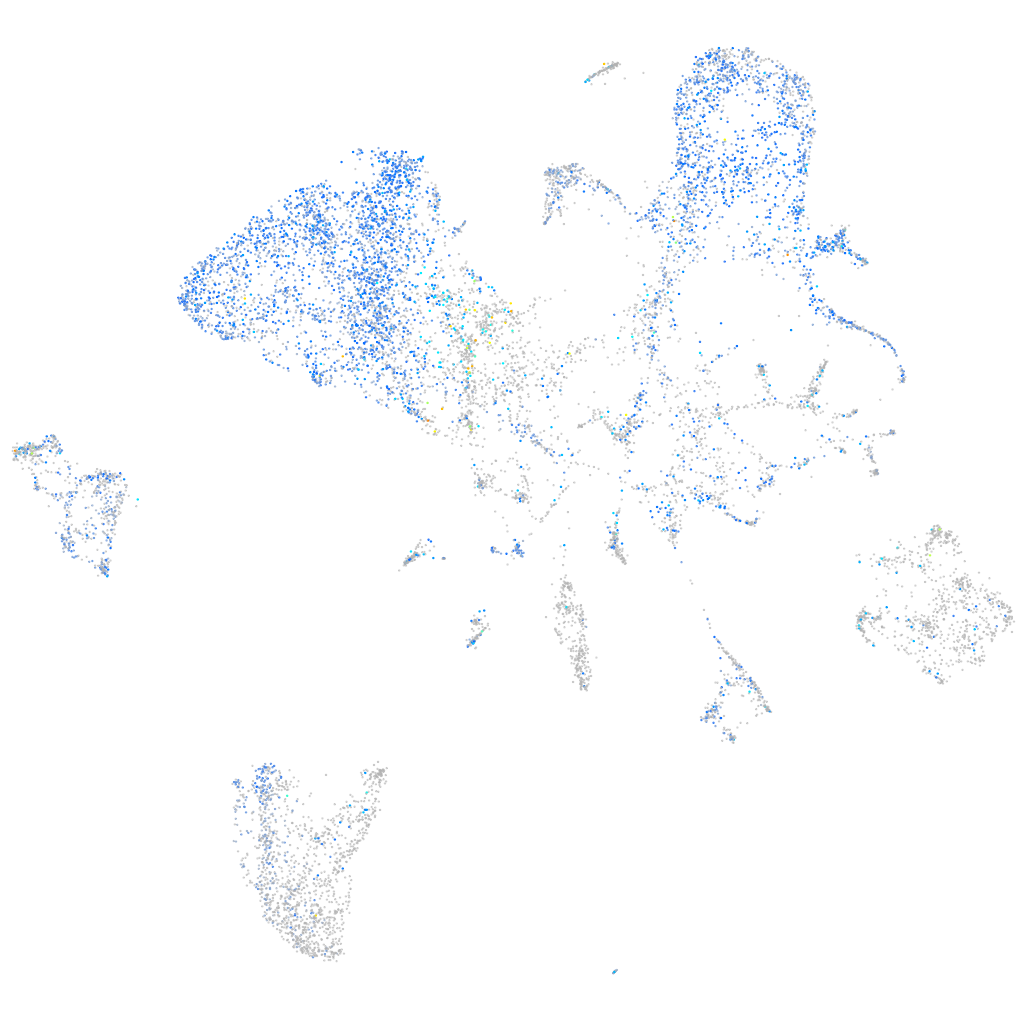
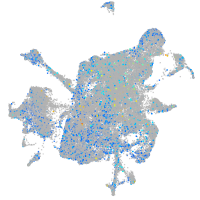
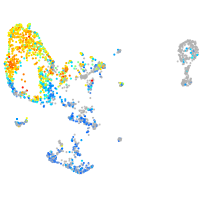
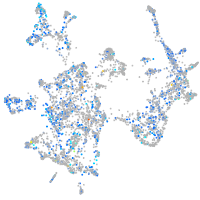
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| scp2a | 0.366 | rtn1a | -0.210 |
| sod1 | 0.366 | ctrl | -0.186 |
| prdx2 | 0.364 | marcksl1b | -0.184 |
| gstt1a | 0.364 | slc38a5b | -0.181 |
| eno3 | 0.359 | ela2l | -0.180 |
| gstr | 0.359 | sycn.2 | -0.180 |
| suclg1 | 0.357 | cpa5 | -0.178 |
| tdo2a | 0.346 | zgc:112160 | -0.178 |
| sult2st2 | 0.345 | ela3l | -0.178 |
| nipsnap3a | 0.345 | ela2 | -0.177 |
| gcshb | 0.343 | zgc:136461 | -0.177 |
| haao | 0.340 | c6ast4 | -0.177 |
| mdh1aa | 0.340 | cpb1 | -0.176 |
| ugt5b4 | 0.336 | ctrb1 | -0.176 |
| apoa4b.1 | 0.335 | CELA1 (1 of many) | -0.175 |
| rdh1 | 0.335 | cpa4 | -0.174 |
| ckba | 0.331 | prss59.1 | -0.174 |
| gpx4a | 0.331 | prss59.2 | -0.173 |
| pgam1a | 0.331 | cel.2 | -0.173 |
| sdr16c5b | 0.330 | cpa1 | -0.172 |
| dhrs9 | 0.329 | si:ch211-240l19.5 | -0.172 |
| mgst1.2 | 0.329 | zgc:92137 | -0.172 |
| acadm | 0.327 | amy2a | -0.172 |
| rmdn1 | 0.326 | erp27 | -0.170 |
| acmsd | 0.325 | cel.1 | -0.170 |
| pgk1 | 0.324 | fep15 | -0.170 |
| gpx1a | 0.324 | apoda.2 | -0.169 |
| srd5a2a | 0.324 | pdia2 | -0.168 |
| phyhd1 | 0.323 | prss1 | -0.166 |
| slc37a4a | 0.323 | si:dkey-14d8.6 | -0.165 |
| sod2 | 0.322 | marcksb | -0.162 |
| atp5mc1 | 0.322 | endou | -0.160 |
| slco1d1 | 0.321 | si:ch211-240l19.6 | -0.157 |
| fdx1 | 0.320 | hmgb3a | -0.155 |
| cat | 0.319 | pla2g1b | -0.154 |