"nuclear factor, interleukin 3 regulated, member 5"
ZFIN 
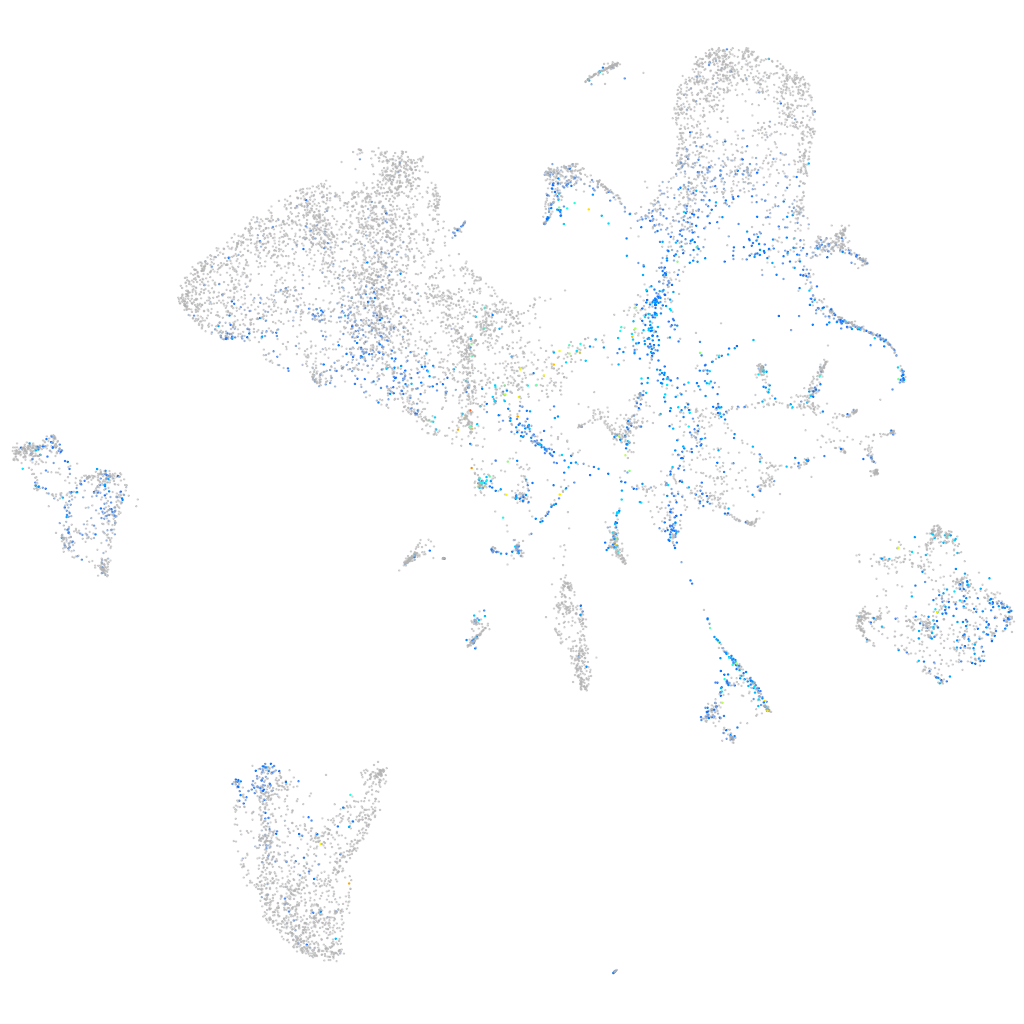
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:ch211-222l21.1 | 0.305 | pnp4b | -0.247 |
| cirbpa | 0.279 | gatm | -0.243 |
| khdrbs1a | 0.278 | gamt | -0.235 |
| hmgn2 | 0.269 | cyp2n13 | -0.208 |
| cirbpb | 0.268 | gapdh | -0.203 |
| serbp1a | 0.268 | bhmt | -0.199 |
| h2afvb | 0.267 | agxtb | -0.199 |
| snrpf | 0.266 | gpx4a | -0.198 |
| ptmab | 0.264 | gda | -0.196 |
| hmga1a | 0.260 | apobb.1 | -0.194 |
| tmsb4x | 0.260 | apoa4b.1 | -0.192 |
| sub1a | 0.259 | afp4 | -0.192 |
| hnrnpa0b | 0.256 | pck1 | -0.190 |
| hnrnpabb | 0.255 | fetub | -0.188 |
| si:ch73-1a9.3 | 0.253 | ucp1 | -0.188 |
| hnrnpaba | 0.252 | abat | -0.188 |
| setb | 0.252 | cyp2ad2 | -0.187 |
| h3f3d | 0.251 | mat1a | -0.187 |
| hmgb2b | 0.250 | rbp4 | -0.185 |
| snrpd1 | 0.250 | ckba | -0.185 |
| col18a1a | 0.247 | mgst1.2 | -0.183 |
| shha | 0.246 | etnppl | -0.183 |
| seta | 0.245 | ces2 | -0.181 |
| ddx39ab | 0.241 | faxdc2 | -0.181 |
| cldnc | 0.240 | fbp1b | -0.176 |
| cfl1 | 0.239 | comtd1 | -0.175 |
| snrpb | 0.238 | zgc:92744 | -0.173 |
| snrpd2 | 0.236 | apoa1b | -0.173 |
| syncrip | 0.236 | ttr | -0.173 |
| ranbp1 | 0.234 | apoc1 | -0.172 |
| eif4a1a | 0.233 | apom | -0.172 |
| prmt1 | 0.232 | fabp10a | -0.171 |
| nucks1a | 0.232 | dgat2 | -0.171 |
| gng5 | 0.231 | scp2a | -0.170 |
| tuba8l4 | 0.231 | zgc:123103 | -0.170 |