nestin
ZFIN 
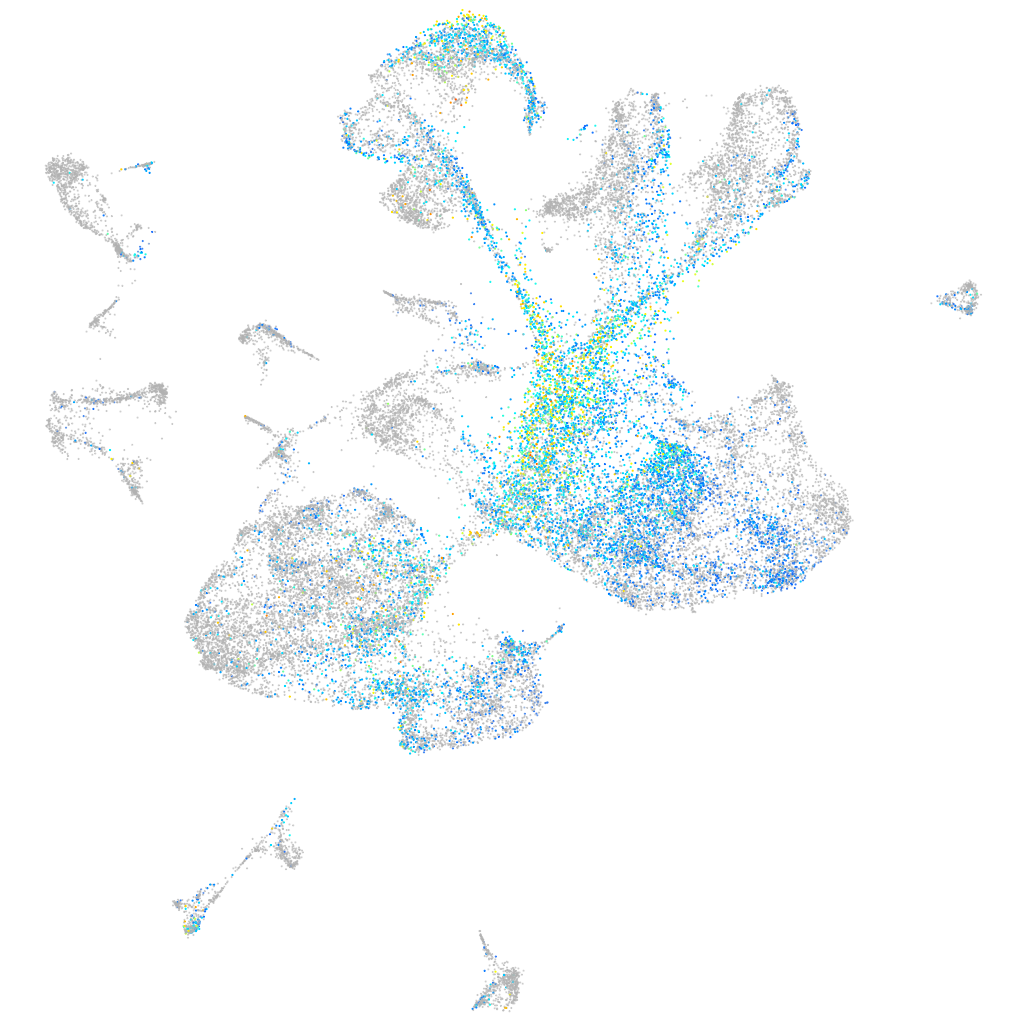
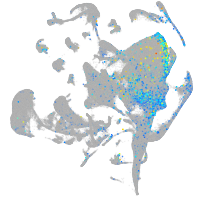
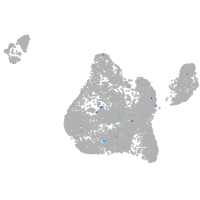
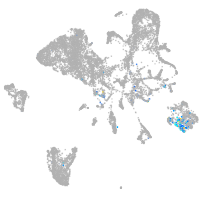
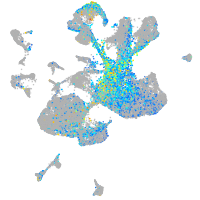
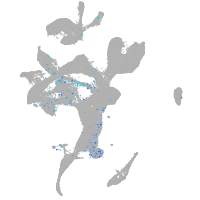
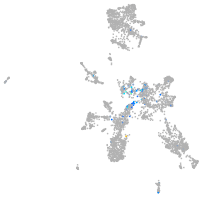
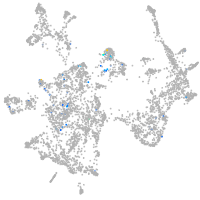
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| LOC798783 | 0.375 | slc1a2b | -0.180 |
| si:ch73-21g5.7 | 0.344 | si:ch211-66e2.5 | -0.158 |
| dlb | 0.342 | glula | -0.157 |
| si:ch211-57n23.4 | 0.342 | cx43 | -0.157 |
| neurod4 | 0.339 | ptn | -0.154 |
| gadd45gb.1 | 0.338 | gapdhs | -0.153 |
| notch1a | 0.327 | acbd7 | -0.144 |
| ebf2 | 0.326 | slc4a4a | -0.143 |
| cep131 | 0.321 | ckbb | -0.142 |
| sox11b | 0.315 | ppap2d | -0.139 |
| her13 | 0.315 | cadm4 | -0.138 |
| LOC101882472 | 0.310 | IGLON5 | -0.137 |
| rtca | 0.309 | cebpd | -0.136 |
| dla | 0.308 | ndrg3a | -0.135 |
| fstl1a | 0.288 | atp1a1b | -0.135 |
| tp53inp2 | 0.286 | hepacama | -0.133 |
| sb:cb81 | 0.281 | cox4i2 | -0.133 |
| lrrn1 | 0.281 | slc3a2a | -0.132 |
| inavaa | 0.276 | anxa13 | -0.132 |
| dld | 0.274 | efhd1 | -0.131 |
| srrm4 | 0.268 | ptgdsb.2 | -0.130 |
| myt1a | 0.263 | igfbp7 | -0.126 |
| cdkn1ca | 0.263 | her9 | -0.126 |
| vim | 0.257 | tpi1b | -0.126 |
| BX530077.1 | 0.256 | atp5if1b | -0.125 |
| sertad2a | 0.255 | nanos1 | -0.125 |
| si:ch211-222l21.1 | 0.250 | mdkb | -0.125 |
| cirbpb | 0.248 | smox | -0.125 |
| sp8b | 0.246 | gpr37l1b | -0.124 |
| XLOC-003689 | 0.246 | mt2 | -0.123 |
| rnf165a | 0.242 | fabp7a | -0.122 |
| fabp3 | 0.241 | dap1b | -0.121 |
| lfng | 0.238 | krt18a.1 | -0.120 |
| hes6 | 0.237 | zgc:153704 | -0.118 |
| acvr2ba | 0.234 | id2a | -0.117 |