NADH:ubiquinone oxidoreductase subunit A3
ZFIN 
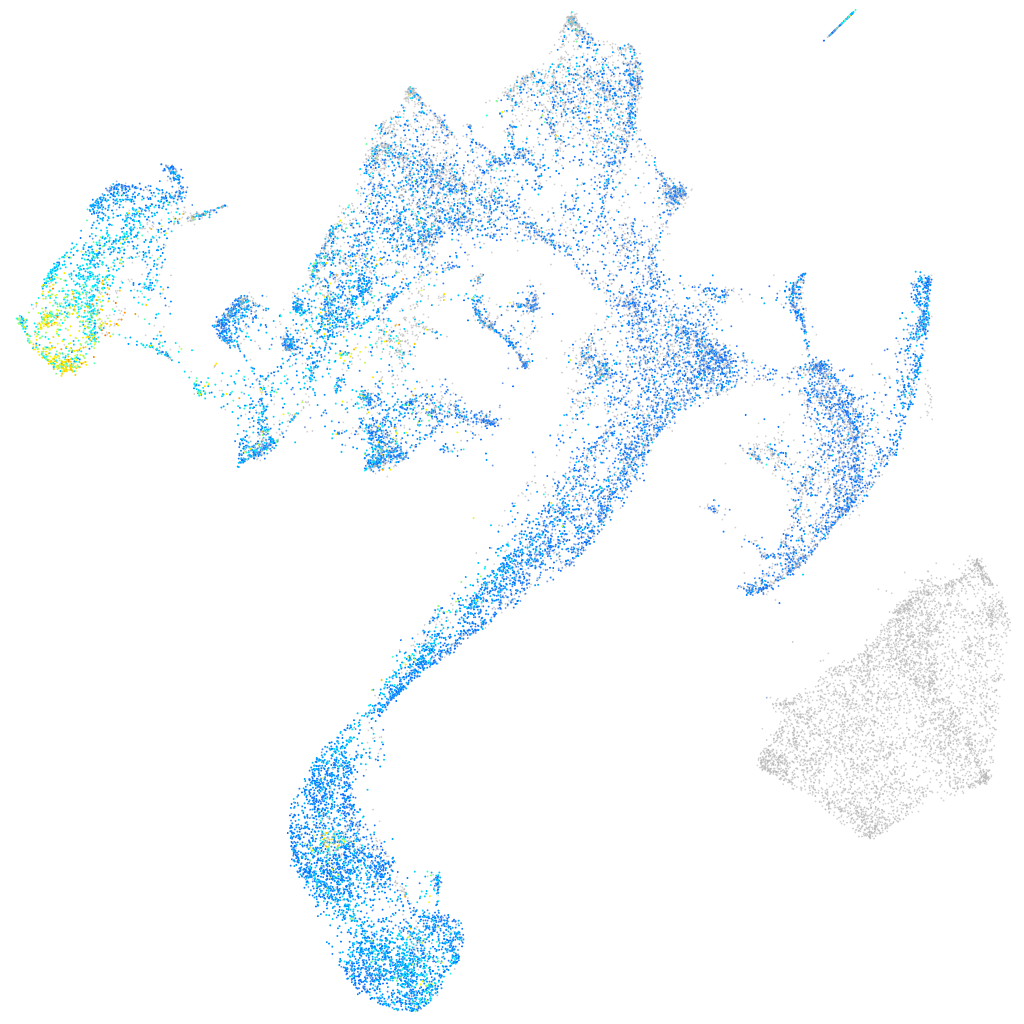
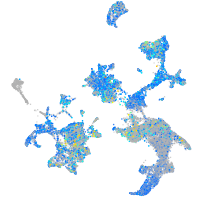
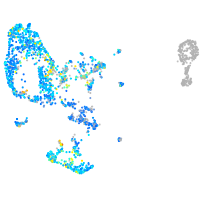
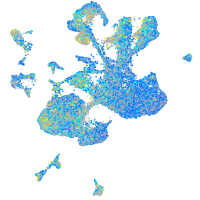
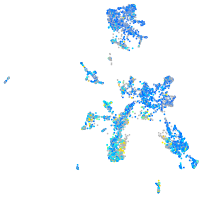
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| atp5meb | 0.547 | si:ch73-1a9.3 | -0.518 |
| atp5if1b | 0.546 | ptmab | -0.490 |
| atp5f1b | 0.537 | pabpc1a | -0.483 |
| aldoab | 0.534 | si:ch211-222l21.1 | -0.475 |
| cox7a2a | 0.530 | hsp90ab1 | -0.474 |
| mdh2 | 0.528 | si:ch73-281n10.2 | -0.464 |
| actc1b | 0.520 | hnrnpaba | -0.463 |
| atp2a1 | 0.519 | hmgb2a | -0.460 |
| atp5l | 0.517 | hmga1a | -0.460 |
| ckmb | 0.516 | h3f3d | -0.459 |
| eno3 | 0.516 | nucks1a | -0.457 |
| ckma | 0.512 | cirbpb | -0.457 |
| gapdh | 0.511 | anp32a | -0.456 |
| nme2b.2 | 0.511 | khdrbs1a | -0.456 |
| atp5mc1 | 0.511 | cirbpa | -0.448 |
| ak1 | 0.510 | cx43.4 | -0.448 |
| atp5mc3b | 0.509 | syncrip | -0.446 |
| ttn.2 | 0.498 | hmgb2b | -0.441 |
| atp5fa1 | 0.496 | seta | -0.438 |
| pabpc4 | 0.494 | acin1a | -0.436 |
| pgam2 | 0.491 | hnrnpabb | -0.435 |
| tmem38a | 0.491 | hnrnpa0b | -0.430 |
| neb | 0.489 | anp32b | -0.426 |
| atp5pf | 0.488 | h2afvb | -0.425 |
| pvalb4 | 0.480 | marcksb | -0.424 |
| ttn.1 | 0.480 | hnrnpa1b | -0.423 |
| cox6a1 | 0.479 | ncl | -0.422 |
| suclg1 | 0.479 | fthl27 | -0.421 |
| mt-atp6 | 0.478 | hmgn7 | -0.418 |
| casq2 | 0.476 | anp32e | -0.411 |
| tpi1b | 0.475 | cbx3a | -0.411 |
| prx | 0.474 | hnrnpub | -0.409 |
| cox8a | 0.474 | hmgn6 | -0.409 |
| srl | 0.472 | ilf3b | -0.405 |
| actn3b | 0.470 | nop58 | -0.402 |