NCK-associated protein 1
ZFIN 
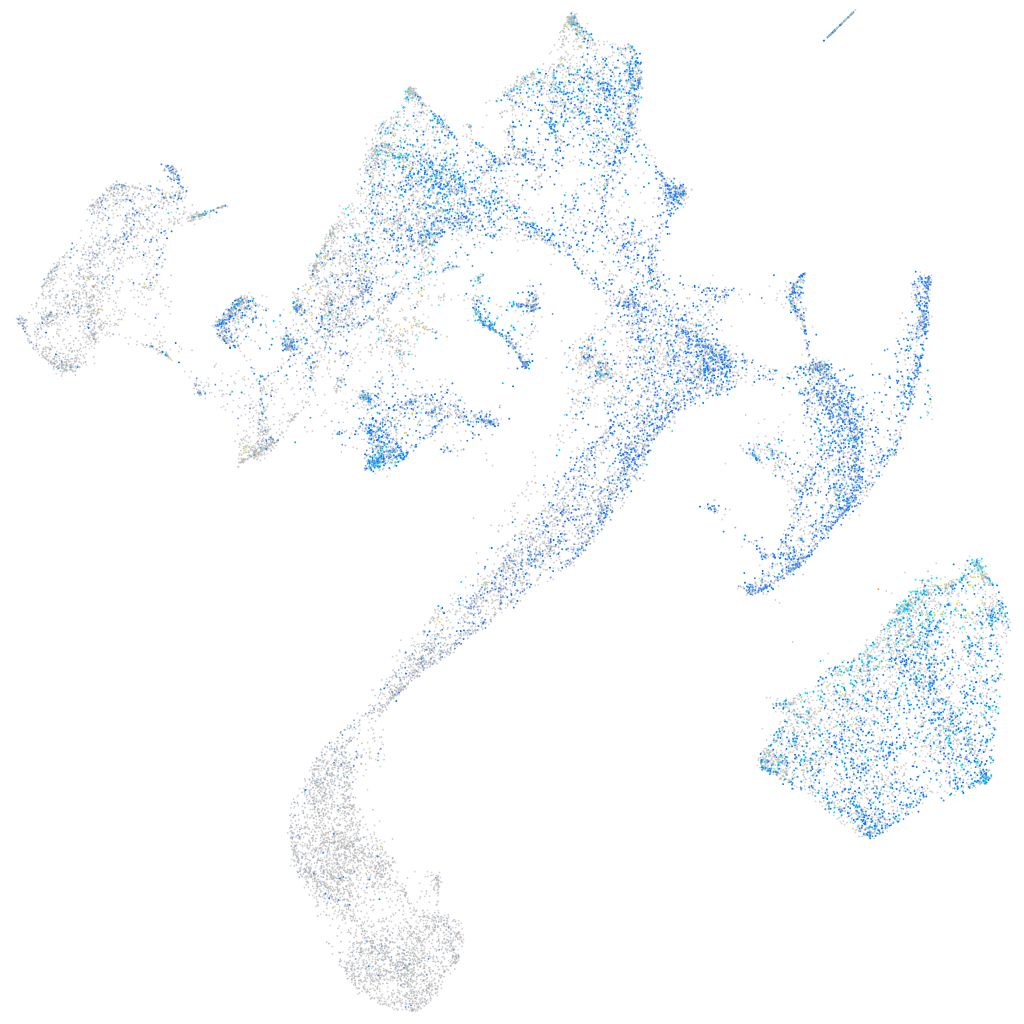
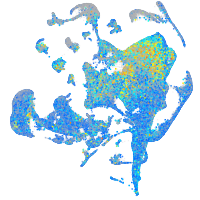
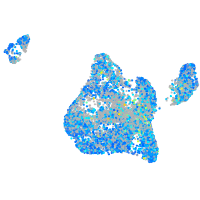
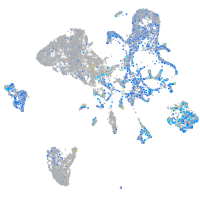
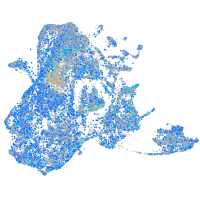
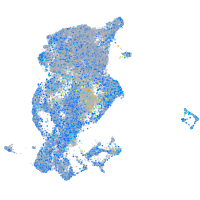
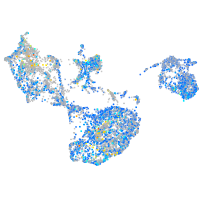
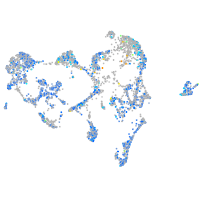
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:ch73-1a9.3 | 0.241 | ckmb | -0.242 |
| hnrnpaba | 0.229 | actc1b | -0.241 |
| cirbpa | 0.227 | ckma | -0.239 |
| h3f3d | 0.225 | atp2a1 | -0.237 |
| si:ch211-222l21.1 | 0.225 | ak1 | -0.235 |
| hmgn6 | 0.223 | tnnc2 | -0.227 |
| ptmab | 0.223 | si:ch73-367p23.2 | -0.225 |
| pabpc1a | 0.223 | aldoab | -0.222 |
| h2afvb | 0.222 | neb | -0.222 |
| hsp90ab1 | 0.221 | actn3a | -0.218 |
| khdrbs1a | 0.220 | ldb3b | -0.218 |
| ubc | 0.220 | tmem38a | -0.218 |
| cirbpb | 0.220 | gapdh | -0.217 |
| hmga1a | 0.217 | mylpfa | -0.216 |
| si:ch73-281n10.2 | 0.213 | pabpc4 | -0.215 |
| nucks1a | 0.212 | mylz3 | -0.213 |
| hmgb2a | 0.211 | nme2b.2 | -0.213 |
| hmgn7 | 0.211 | myom1a | -0.212 |
| hmgb2b | 0.208 | actn3b | -0.212 |
| hnrnpa0b | 0.208 | tnnt3a | -0.212 |
| syncrip | 0.207 | ttn.2 | -0.212 |
| cfl1 | 0.205 | smyd1a | -0.209 |
| anp32a | 0.204 | acta1b | -0.209 |
| si:ch211-288g17.3 | 0.204 | hhatla | -0.208 |
| hnrnpabb | 0.202 | si:ch211-266g18.10 | -0.208 |
| fthl27 | 0.201 | pgam2 | -0.208 |
| hmgn2 | 0.200 | ank1a | -0.206 |
| acin1a | 0.199 | CABZ01078594.1 | -0.206 |
| seta | 0.197 | tpma | -0.205 |
| cx43.4 | 0.194 | tmod4 | -0.205 |
| ywhaqb | 0.192 | pvalb1 | -0.203 |
| hdac1 | 0.192 | myl1 | -0.202 |
| cbx3a | 0.192 | pvalb2 | -0.202 |
| pnrc2 | 0.191 | eno3 | -0.202 |
| hp1bp3 | 0.191 | gamt | -0.201 |