"non-SMC condensin II complex, subunit G2"
ZFIN 
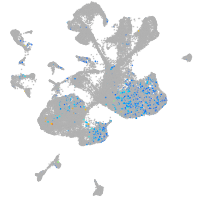
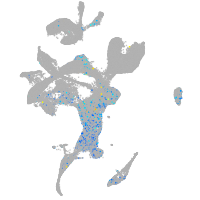
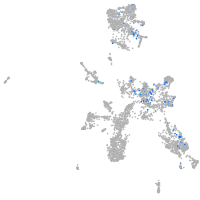
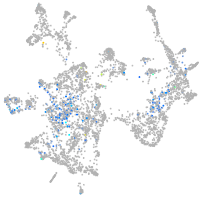
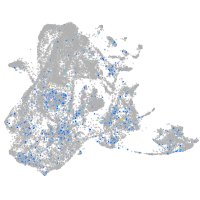
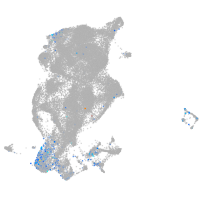
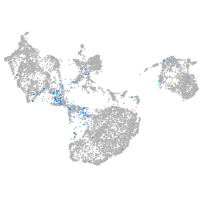
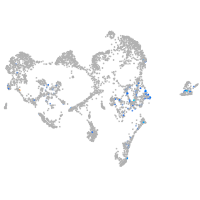
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| esr2a | 0.357 | dap1b | -0.068 |
| smc2 | 0.286 | cox4i2 | -0.054 |
| mki67 | 0.281 | actb2 | -0.053 |
| top2a | 0.280 | zgc:162730 | -0.052 |
| zgc:110425 | 0.250 | tpm1 | -0.051 |
| cenpe | 0.247 | eef1da | -0.051 |
| mibp | 0.246 | tob1b | -0.050 |
| ggact.1 | 0.243 | tmsb4x | -0.049 |
| kif15 | 0.237 | myl9a | -0.049 |
| rrm1 | 0.236 | ier2b | -0.048 |
| kif22 | 0.236 | fn1b | -0.048 |
| CABZ01072848.1 | 0.236 | acta2 | -0.048 |
| cks1b | 0.233 | cebpd | -0.048 |
| gpr186 | 0.233 | gabarapb | -0.047 |
| ncaph2 | 0.232 | rsl24d1 | -0.047 |
| si:dkey-261m9.17 | 0.231 | tagln | -0.047 |
| ccna2 | 0.228 | apoa1b | -0.046 |
| aurkb | 0.225 | ppdpfb | -0.046 |
| nuf2 | 0.225 | rbp4 | -0.045 |
| plk1 | 0.221 | lxn | -0.045 |
| zgc:110540 | 0.218 | apoa2 | -0.044 |
| zgc:153405 | 0.217 | ifitm1 | -0.044 |
| CABZ01005379.1 | 0.217 | ak1 | -0.044 |
| nsl1 | 0.216 | prss59.2 | -0.043 |
| ncapd2 | 0.215 | sesn1 | -0.043 |
| ercc6l | 0.214 | mfap2 | -0.042 |
| cenph | 0.213 | sparc | -0.042 |
| chtf18 | 0.210 | lbh | -0.042 |
| dut | 0.209 | mxra8b | -0.042 |
| dlgap5 | 0.209 | pkig | -0.041 |
| gltpb | 0.209 | iscub | -0.041 |
| ncapd3 | 0.208 | krt8 | -0.041 |
| trip13 | 0.208 | col4a1 | -0.039 |
| si:ch211-266i6.3 | 0.207 | cd248a | -0.039 |
| ncapg | 0.206 | slc25a5 | -0.039 |