neural cell adhesion molecule 1a
ZFIN 
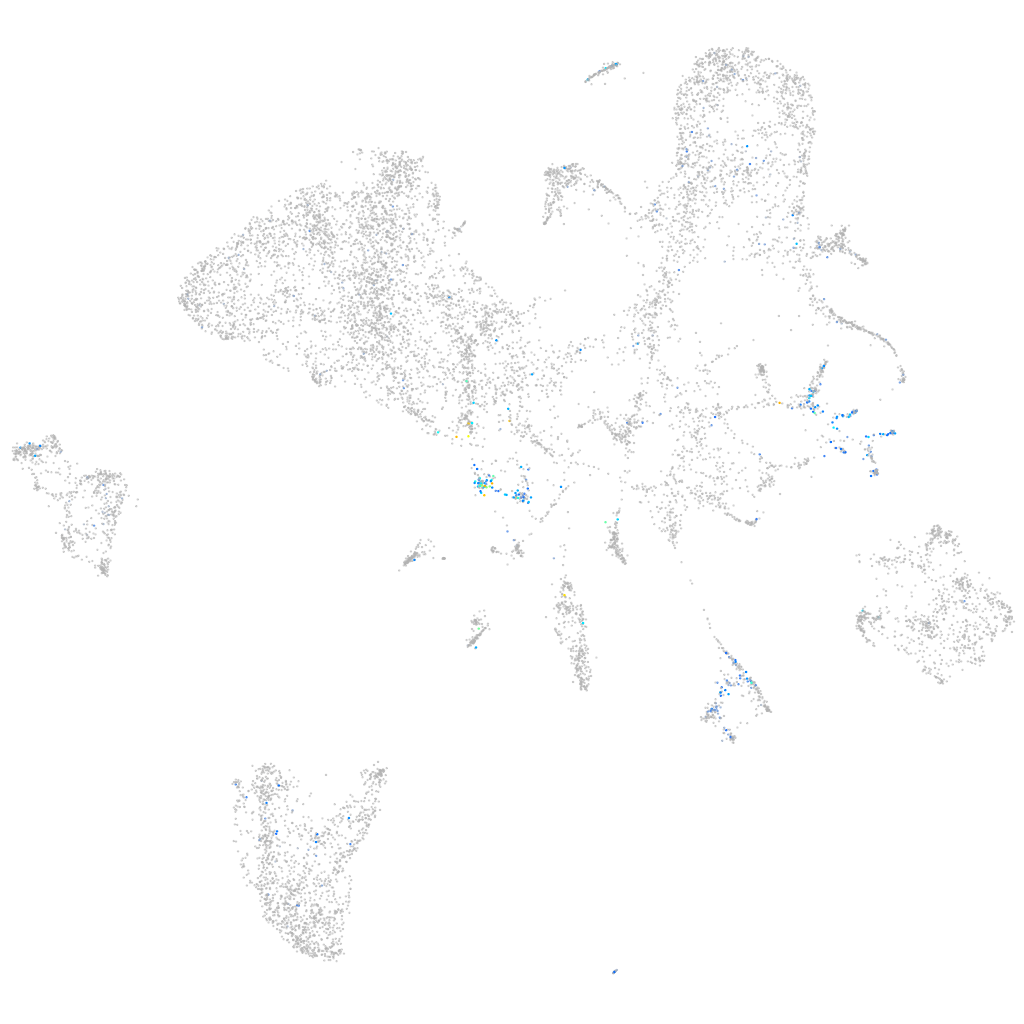
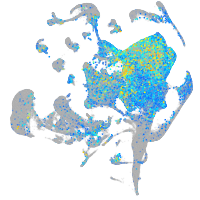
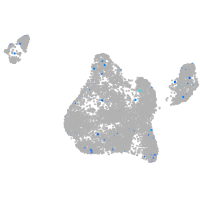
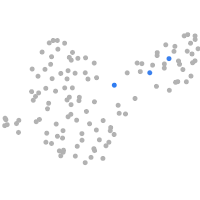
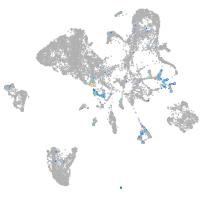
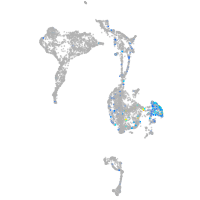
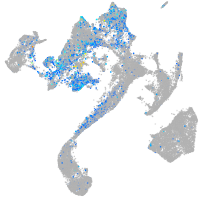
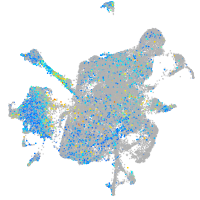
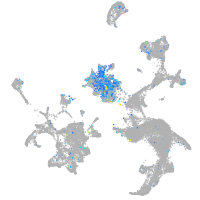
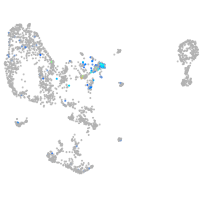
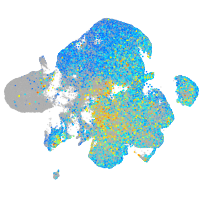
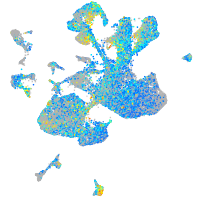
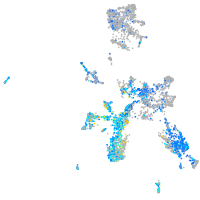
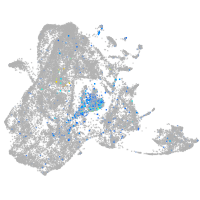
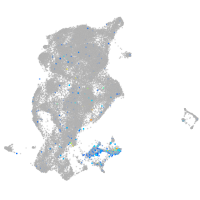
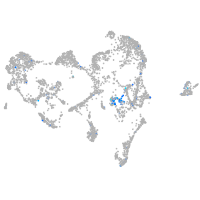
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| elavl3 | 0.310 | aldob | -0.107 |
| stmn1b | 0.304 | gapdh | -0.106 |
| tuba1c | 0.278 | gamt | -0.093 |
| nova2 | 0.264 | gstr | -0.092 |
| myt1b | 0.264 | gstt1a | -0.090 |
| elavl4 | 0.256 | aldh7a1 | -0.089 |
| gpm6aa | 0.256 | aldh6a1 | -0.089 |
| dpysl3 | 0.244 | pnp4b | -0.089 |
| tuba1a | 0.243 | fbp1b | -0.089 |
| LOC100537384 | 0.242 | gatm | -0.088 |
| rnasekb | 0.240 | scp2a | -0.087 |
| ebf2 | 0.236 | mat1a | -0.087 |
| mapk10 | 0.236 | eno3 | -0.086 |
| gng3 | 0.231 | ahcy | -0.085 |
| lhx2b | 0.225 | agxta | -0.084 |
| si:dkey-153k10.9 | 0.225 | glud1b | -0.084 |
| cadm3 | 0.222 | rdh1 | -0.082 |
| CU467822.1 | 0.218 | ttc36 | -0.082 |
| slc35g2b | 0.215 | ckba | -0.081 |
| scn8aa | 0.214 | gnmt | -0.081 |
| vamp2 | 0.214 | agxtb | -0.080 |
| mdkb | 0.214 | etnppl | -0.080 |
| tmsb | 0.213 | serpinf2b | -0.080 |
| myt1a | 0.212 | igfbp2a | -0.080 |
| neurod1 | 0.212 | nipsnap3a | -0.080 |
| sncb | 0.211 | aqp12 | -0.080 |
| tspan7b | 0.211 | abat | -0.079 |
| vat1 | 0.210 | uox | -0.079 |
| cnrip1a | 0.210 | pck2 | -0.079 |
| mtus2a | 0.210 | tdo2a | -0.079 |
| st8sia5 | 0.208 | cpn1 | -0.079 |
| gnao1a | 0.208 | suclg2 | -0.078 |
| si:dkeyp-75h12.5 | 0.208 | mgst1.2 | -0.078 |
| celf4 | 0.204 | g6pca.2 | -0.078 |
| rtn1b | 0.203 | cx32.3 | -0.078 |