N-acetylneuraminic acid synthase a
ZFIN 
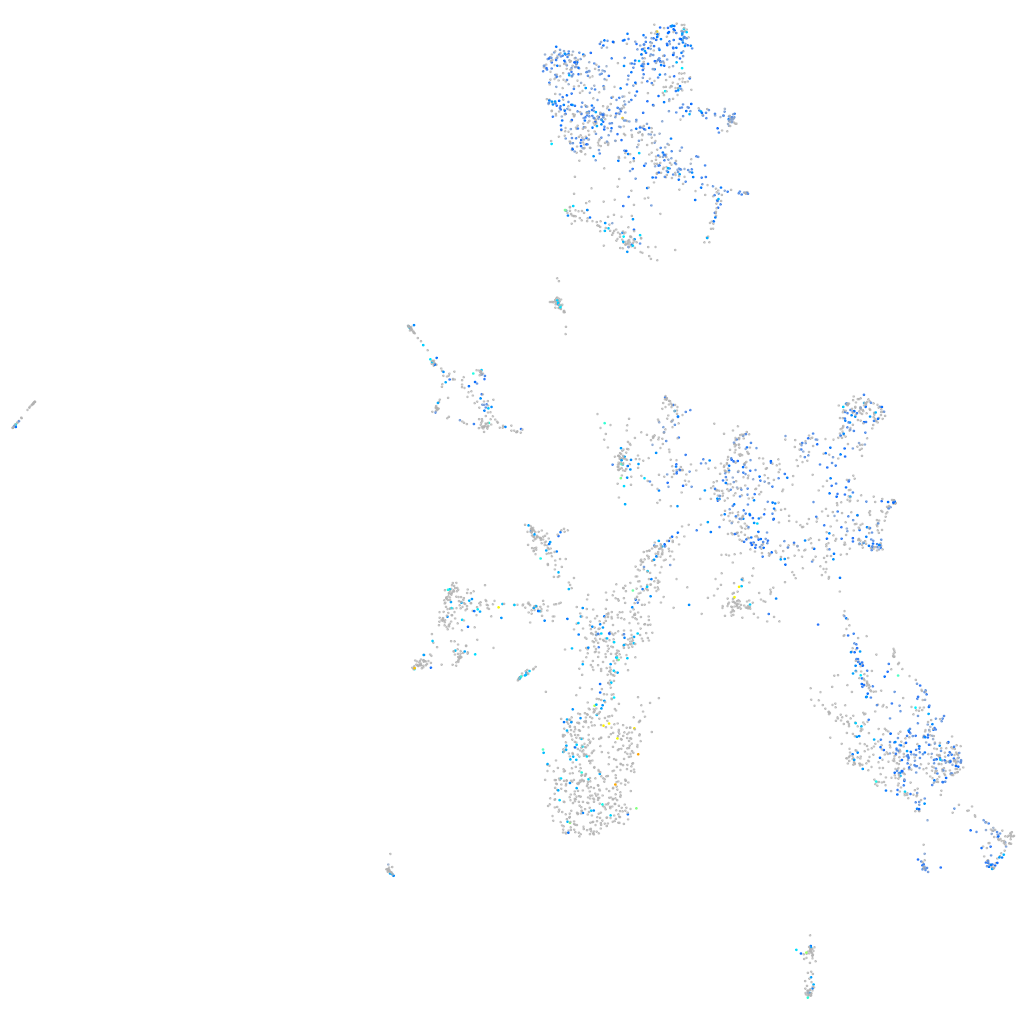
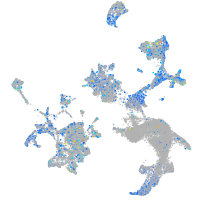
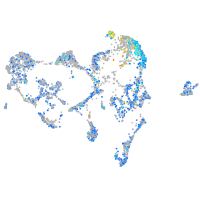
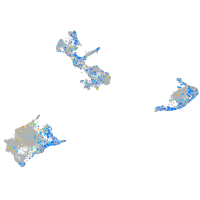
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:ch211-157c3.4 | 0.167 | NC-002333.4 | -0.095 |
| ppl | 0.166 | cdc25d | -0.074 |
| si:dkey-19b23.12 | 0.160 | kcnma1a | -0.073 |
| s100v2 | 0.155 | pim3 | -0.071 |
| s100v1 | 0.153 | si:ch211-270n8.4 | -0.070 |
| capns1a | 0.151 | atf5a | -0.069 |
| tubb4b | 0.147 | fabp10b | -0.069 |
| ponzr5 | 0.147 | cnga4 | -0.069 |
| s100w | 0.147 | nrsn1 | -0.068 |
| sult6b1 | 0.146 | abhd2b | -0.068 |
| si:dkey-87o1.2 | 0.145 | nxnl2 | -0.067 |
| krt92 | 0.145 | pvalb5 | -0.066 |
| GCA | 0.145 | glula | -0.066 |
| scel | 0.144 | elavl3 | -0.065 |
| krt1-19d | 0.144 | tspan2a | -0.064 |
| anxa3a | 0.143 | CR383676.1 | -0.064 |
| si:ch73-347e22.8 | 0.143 | tshz2 | -0.064 |
| agr1 | 0.142 | ttc9b | -0.064 |
| cd9b | 0.142 | scg2b | -0.063 |
| dspa | 0.139 | atp2b1a | -0.063 |
| si:ch73-52f15.5 | 0.138 | NC-002333.17 | -0.062 |
| pitx1 | 0.138 | lhx2b | -0.061 |
| b3gnt7l | 0.138 | myl13 | -0.061 |
| capn9 | 0.137 | serpini1 | -0.061 |
| egr1 | 0.137 | ptp4a3 | -0.061 |
| zgc:153665 | 0.135 | rarres3l | -0.060 |
| pycard | 0.134 | ptbp2b | -0.060 |
| gmds | 0.133 | mtbl | -0.060 |
| gbgt1l3 | 0.133 | LOC103910744 | -0.059 |
| arpc2 | 0.133 | cyfip2 | -0.059 |
| actb1 | 0.132 | XLOC-024343 | -0.059 |
| pfn2 | 0.132 | nsg2 | -0.058 |
| mcl1a | 0.131 | fstl5 | -0.057 |
| psma1 | 0.131 | sprn2 | -0.057 |
| mgst2 | 0.131 | myo5aa | -0.057 |