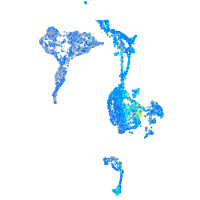
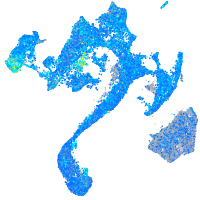
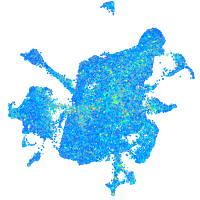
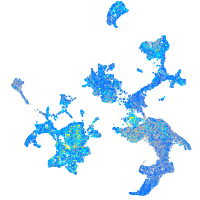
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| NC-002333.4 | 0.624 | si:dkey-33c14.3 | -0.304 |
| mt-co1 | 0.421 | cxl34b.11 | -0.293 |
| mt-co2 | 0.407 | zgc:136930 | -0.286 |
| CABZ01072614.1 | 0.396 | ahcy | -0.285 |
| COX3 | 0.371 | krt91 | -0.274 |
| mt-cyb | 0.351 | si:dkey-183i3.5 | -0.272 |
| zgc:158463 | 0.314 | krt5 | -0.270 |
| LOC110439372 | 0.307 | epgn | -0.269 |
| CR383676.1 | 0.306 | zgc:77439 | -0.264 |
| mt-nd3 | 0.272 | krtt1c19e | -0.255 |
| mt-nd4 | 0.235 | lgals1l1 | -0.249 |
| dag1 | 0.231 | rbp4 | -0.234 |
| mt-nd2 | 0.229 | eef1da | -0.233 |
| cdh1 | 0.228 | ppifb | -0.232 |
| mt-atp6 | 0.226 | glo1 | -0.225 |
| akap12b | 0.226 | eef1b2 | -0.222 |
| AL935186.9 | 0.217 | gapdhs | -0.218 |
| CABZ01075068.1 | 0.217 | eif4ebp3 | -0.218 |
| asph | 0.215 | mgarp | -0.213 |
| lamb1a | 0.213 | ucp2 | -0.212 |
| mt-nd1 | 0.211 | romo1 | -0.210 |
| nid2a | 0.211 | pnp5a | -0.206 |
| bcam | 0.209 | rpl7 | -0.204 |
| col14a1a | 0.204 | rpl19 | -0.204 |
| fkbp9 | 0.201 | prelid3b | -0.202 |
| fbn2b | 0.201 | si:ch211-76l23.4 | -0.199 |
| arid3c | 0.192 | capns1a | -0.196 |
| btg1 | 0.192 | cebpd | -0.195 |
| igf2bp1 | 0.188 | mylipb | -0.192 |
| mt-nd5 | 0.188 | anxa1a | -0.191 |
| acin1a | 0.183 | aldh9a1a.1 | -0.189 |
| rrbp1b | 0.182 | cebpb | -0.186 |
| p3h1 | 0.182 | ptgdsb.1 | -0.185 |
| p4ha1b | 0.181 | wu:fa03e10 | -0.184 |
| lsp1 | 0.178 | cdo1 | -0.182 |