"myosin, light polypeptide 3, skeletal muscle"
ZFIN 
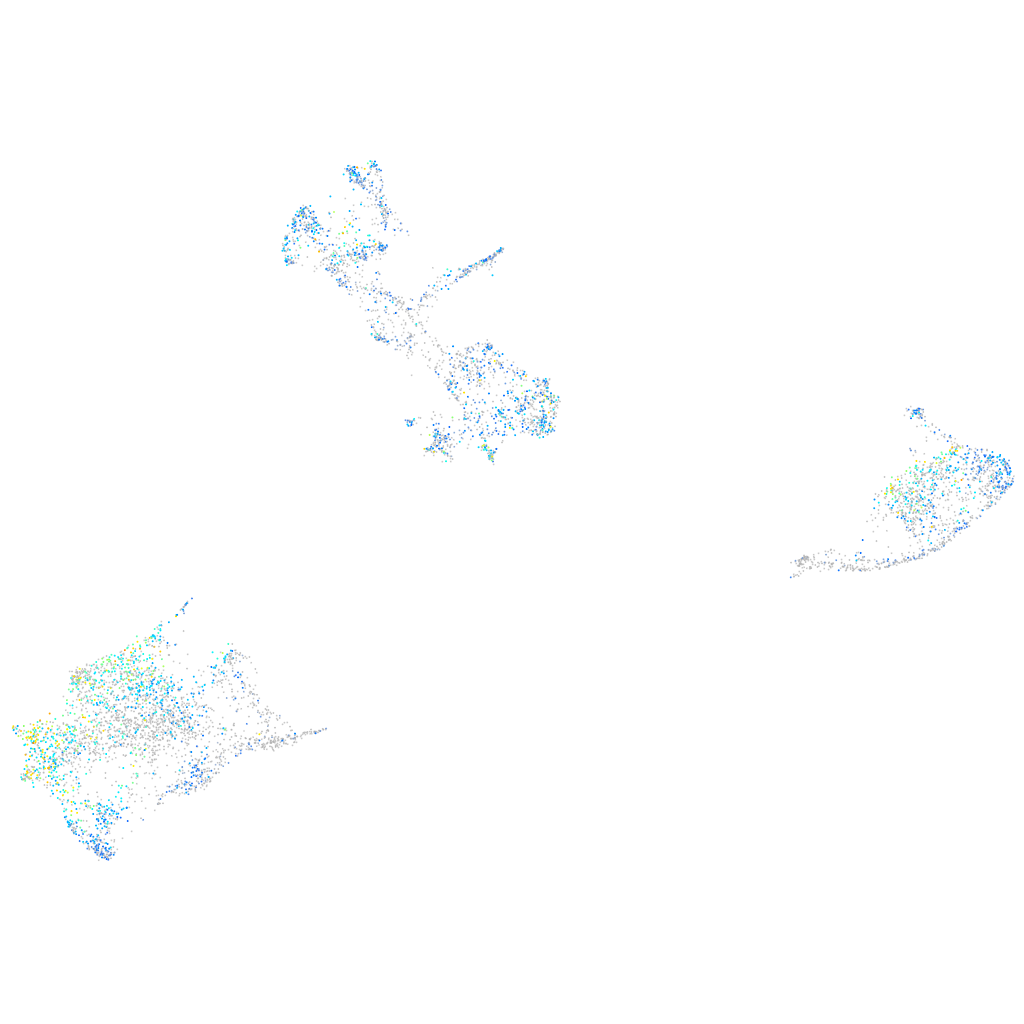
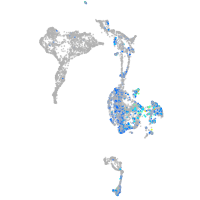
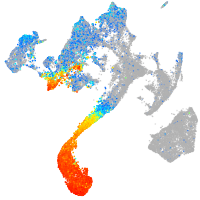
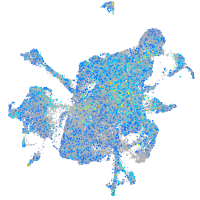
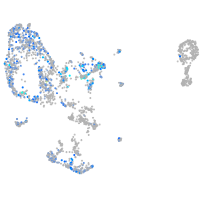
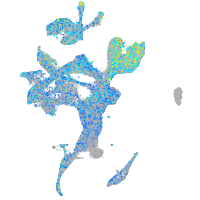
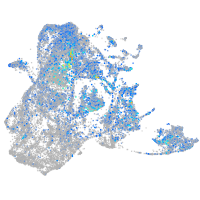
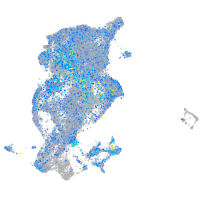
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| pvalb2 | 0.432 | cfl1 | -0.159 |
| pvalb1 | 0.418 | ptges3b | -0.158 |
| actc1b | 0.397 | hsp90ab1 | -0.150 |
| mylpfa | 0.341 | rps12 | -0.147 |
| tnnt3b | 0.298 | tcp1 | -0.147 |
| tnni2a.4 | 0.278 | ssr4 | -0.146 |
| tpma | 0.255 | aldoaa | -0.144 |
| myhz1.1 | 0.245 | cox8a | -0.143 |
| mylpfb | 0.245 | sec61b | -0.143 |
| nme2b.2 | 0.242 | serbp1a | -0.142 |
| cyt1 | 0.230 | rplp2l | -0.142 |
| apoa2 | 0.226 | hmga1a | -0.140 |
| ttn.1 | 0.226 | rps26 | -0.140 |
| ckma | 0.223 | snrpe | -0.138 |
| ckmb | 0.216 | eif2s2 | -0.137 |
| acta1b | 0.215 | hnrnpa0l | -0.136 |
| krt4 | 0.208 | rpl13a | -0.136 |
| apoa1b | 0.206 | npm1a | -0.134 |
| myl1 | 0.201 | snrpg | -0.134 |
| ttn.2 | 0.193 | rpl17 | -0.133 |
| tnnc2 | 0.172 | lman2 | -0.133 |
| krt17 | 0.165 | cox7a2a | -0.133 |
| hbbe1.3 | 0.164 | tmem258 | -0.132 |
| myhz1.2 | 0.158 | psma5 | -0.131 |
| krtt1c19e | 0.155 | slc25a3b | -0.131 |
| icn | 0.154 | pfdn4 | -0.131 |
| prss59.2 | 0.153 | calr3b | -0.131 |
| hbae3 | 0.150 | fkbp2 | -0.131 |
| myhz2 | 0.147 | bzw1a | -0.131 |
| krt91 | 0.147 | hspe1 | -0.130 |
| atp2a1 | 0.146 | dad1 | -0.130 |
| hbae1.1 | 0.144 | cct2 | -0.130 |
| ak1 | 0.141 | xbp1 | -0.130 |
| gapdh | 0.137 | hspd1 | -0.129 |
| prss1 | 0.137 | eif5a2 | -0.129 |