"myosin, light chain 13"
ZFIN 
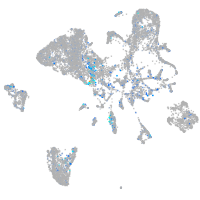
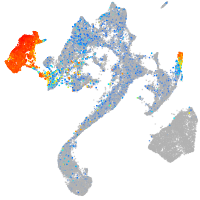
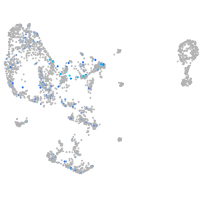
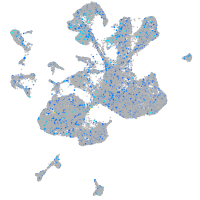
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| actc1b | 0.180 | wu:fb18f06 | -0.074 |
| pvalb2 | 0.179 | rpl32 | -0.070 |
| pvalb4 | 0.177 | rpl23a | -0.069 |
| pvalb1 | 0.172 | lgals3b | -0.068 |
| mylz3 | 0.158 | wu:fa03e10 | -0.067 |
| mylpfa | 0.150 | eef2b | -0.066 |
| apoa2 | 0.147 | cd9b | -0.064 |
| myl10 | 0.138 | uba52 | -0.064 |
| ckmb | 0.129 | prr13 | -0.064 |
| nme2b.2 | 0.126 | cfl1l | -0.062 |
| LOC101885014 | 0.122 | rps18 | -0.062 |
| ckma | 0.122 | rpl27a | -0.062 |
| prss59.2 | 0.120 | capgb | -0.062 |
| crygmx | 0.120 | rps12 | -0.062 |
| mylpfb | 0.117 | rpl30 | -0.061 |
| fabp1b.1 | 0.115 | mtfp1 | -0.061 |
| tnni4b.1 | 0.113 | anxa1a | -0.061 |
| hbae3 | 0.112 | rpl18a | -0.061 |
| gapdh | 0.112 | cldn1 | -0.061 |
| atp2a1 | 0.111 | perp | -0.061 |
| tnni2a.4 | 0.108 | anxa2a | -0.061 |
| ttn.2 | 0.106 | krt5 | -0.060 |
| apoa1b | 0.105 | ahnak | -0.060 |
| trim101 | 0.102 | dhrs13a.2 | -0.060 |
| crygm2d10 | 0.101 | abracl | -0.060 |
| hbae1.1 | 0.100 | pfn1 | -0.059 |
| si:dkey-4p15.5 | 0.099 | abcb5 | -0.059 |
| myl1 | 0.098 | sh3gl1a | -0.059 |
| si:cabz01036022.1 | 0.098 | foxq1a | -0.059 |
| LOC100147871 | 0.095 | tmem176l.4 | -0.059 |
| ttn.1 | 0.095 | rpl31 | -0.059 |
| lgals2b | 0.094 | pabpc1a | -0.058 |
| BX640512.3 | 0.094 | jupa | -0.058 |
| tcnba | 0.094 | rpl34 | -0.058 |
| zgc:136461 | 0.093 | s100a10b | -0.057 |