"myosin, heavy polypeptide 1.1, skeletal muscle"
ZFIN 
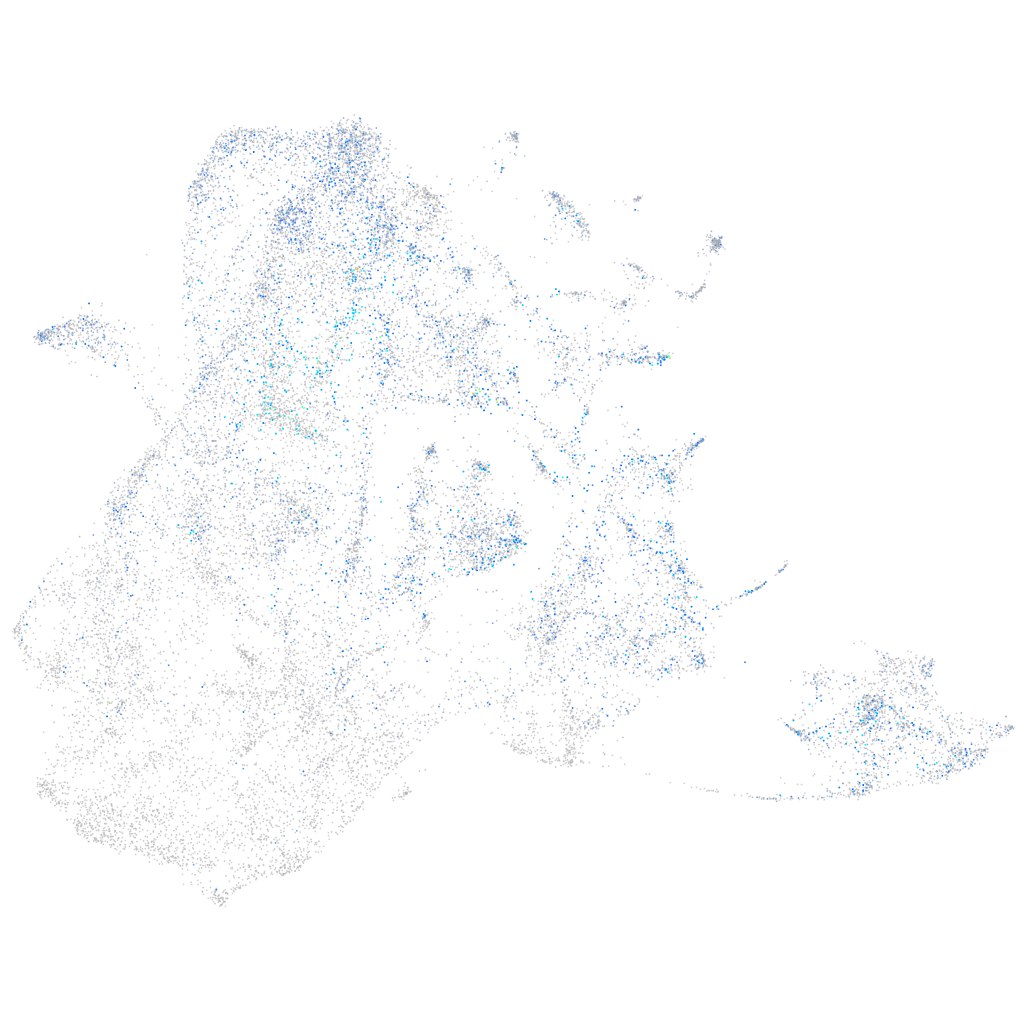
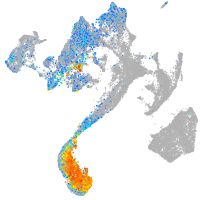
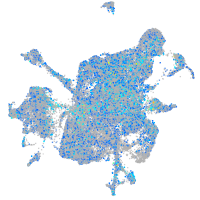
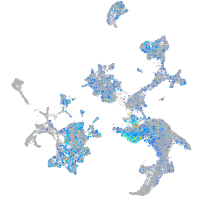
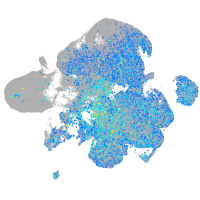
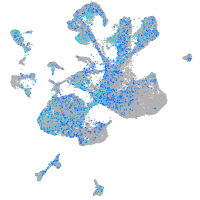
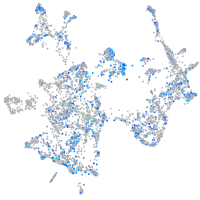
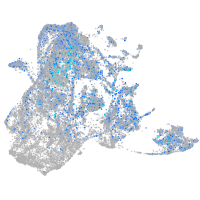
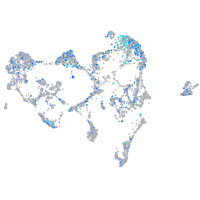
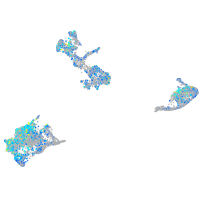
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| pvalb2 | 0.344 | ssr3 | -0.172 |
| pvalb1 | 0.343 | tpm4a | -0.169 |
| mylpfa | 0.320 | ssr2 | -0.158 |
| actc1b | 0.318 | ssr4 | -0.156 |
| mylz3 | 0.260 | dad1 | -0.152 |
| tnni2a.4 | 0.226 | selenof | -0.151 |
| tnnt3b | 0.212 | aldoaa | -0.147 |
| tpma | 0.205 | snrpb | -0.145 |
| mylpfb | 0.203 | lin28a | -0.145 |
| ckma | 0.202 | arf5 | -0.144 |
| myhz1.2 | 0.191 | igf2bp1 | -0.142 |
| apoa2 | 0.190 | tmed2 | -0.142 |
| nme2b.2 | 0.176 | CABZ01075068.1 | -0.142 |
| apoa1b | 0.165 | nid2a | -0.142 |
| hbbe1.3 | 0.164 | cx43.4 | -0.141 |
| ttn.1 | 0.159 | ywhaqa | -0.140 |
| ckmb | 0.158 | akap12b | -0.140 |
| ttn.2 | 0.158 | hmga1a | -0.139 |
| prss59.2 | 0.151 | tspan7 | -0.138 |
| hbae1.1 | 0.149 | crtap | -0.137 |
| myhz2 | 0.149 | tram1 | -0.137 |
| tnnc2 | 0.148 | myl6 | -0.135 |
| hbae3 | 0.146 | cnn2 | -0.134 |
| myl1 | 0.143 | fkbp9 | -0.134 |
| myhz1.3 | 0.142 | ptges3b | -0.133 |
| atp2a1 | 0.138 | p3h1 | -0.133 |
| acta1b | 0.134 | BX927258.1 | -0.133 |
| CELA1 (1 of many) | 0.131 | sept2 | -0.132 |
| prss1 | 0.128 | ddost | -0.131 |
| pvalb4 | 0.123 | snrpd2 | -0.131 |
| zgc:158463 | 0.123 | sec61b | -0.129 |
| hbbe2 | 0.122 | trim71 | -0.129 |
| slc25a4 | 0.114 | hspb1 | -0.128 |
| si:ch73-367p23.2 | 0.112 | hnrnpaba | -0.128 |
| ela2l | 0.110 | fermt1 | -0.128 |