"myosin, heavy chain 14, non-muscle"
ZFIN 
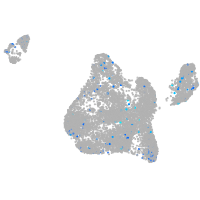
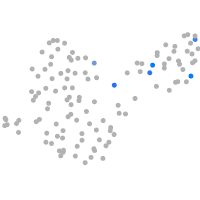
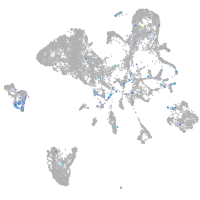
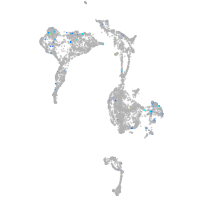
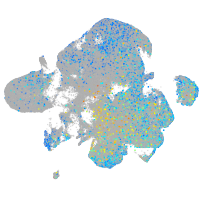
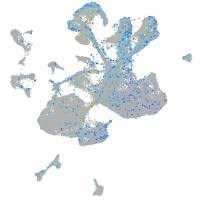
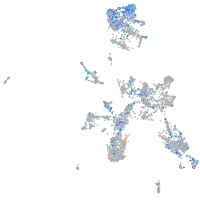
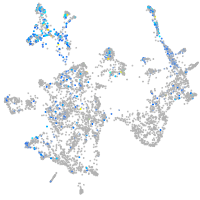
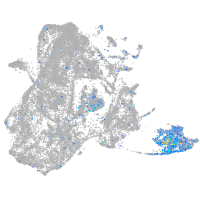
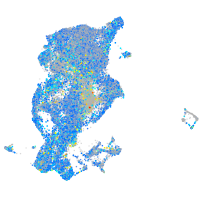
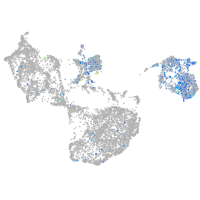
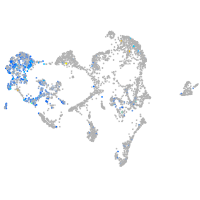
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| gzma | 0.143 | si:dkey-151g10.6 | -0.096 |
| spock3 | 0.136 | paics | -0.085 |
| stx1b | 0.128 | CABZ01021592.1 | -0.073 |
| rtn1a | 0.127 | uraha | -0.071 |
| ckbb | 0.120 | mdh1aa | -0.069 |
| prkar1b | 0.119 | si:dkey-251i10.2 | -0.066 |
| CR383676.1 | 0.118 | rplp1 | -0.065 |
| ppp1r9a | 0.118 | impdh1b | -0.065 |
| gnao1a | 0.118 | aldob | -0.060 |
| SPAG9 | 0.118 | cyb5a | -0.060 |
| ppfia2 | 0.117 | prdx1 | -0.059 |
| dpysl3 | 0.115 | tmem130 | -0.058 |
| BX469918.1 | 0.115 | rplp0 | -0.056 |
| XLOC-005982 | 0.114 | gpd1b | -0.056 |
| PRKAR2B | 0.113 | atic | -0.053 |
| sypa | 0.112 | rps29 | -0.052 |
| gpm6aa | 0.112 | pts | -0.051 |
| gatad2b | 0.112 | rps12 | -0.050 |
| dao.1 | 0.112 | rps5 | -0.050 |
| atp6v0cb | 0.111 | glulb | -0.050 |
| mc1r | 0.111 | actb2 | -0.050 |
| zgc:194665 | 0.111 | prtfdc1 | -0.048 |
| gng3 | 0.111 | rpl19 | -0.048 |
| si:ch73-287m6.1 | 0.111 | gch2 | -0.048 |
| elavl4 | 0.110 | rps23 | -0.047 |
| kmt2cb | 0.108 | rps7 | -0.047 |
| nova2 | 0.107 | rps3a | -0.046 |
| parp12b | 0.107 | akr1b1 | -0.045 |
| elavl3 | 0.107 | zgc:114188 | -0.044 |
| SEMA4F | 0.106 | rplp2l | -0.044 |
| rap1gap | 0.106 | prdx5 | -0.043 |
| rusc1 | 0.106 | iah1 | -0.043 |
| rac3b | 0.106 | phyhd1 | -0.043 |
| slc6a15 | 0.105 | rbp4l | -0.043 |
| cacna1aa | 0.104 | mibp | -0.041 |