MAX dimerization protein 4
ZFIN 
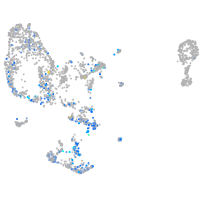
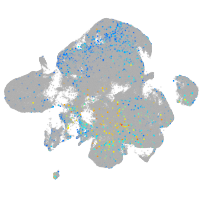
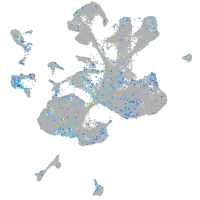
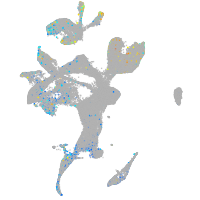
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| aldocb | 0.083 | si:ch211-222l21.1 | -0.095 |
| sncgb | 0.080 | hmga1a | -0.086 |
| vsnl1b | 0.078 | hnrnpabb | -0.081 |
| nupr1a | 0.078 | khdrbs1a | -0.075 |
| eno1a | 0.076 | ptmab | -0.070 |
| atp2b2 | 0.075 | hnrnpaba | -0.069 |
| ddit3 | 0.075 | cirbpa | -0.069 |
| eno2 | 0.075 | hmgb2b | -0.068 |
| snap25b | 0.073 | hnrnpa0b | -0.068 |
| gapdhs | 0.073 | seta | -0.067 |
| slc6a1a | 0.072 | snrpf | -0.065 |
| calm1b | 0.071 | rbbp4 | -0.063 |
| ndrg3a | 0.071 | top1l | -0.062 |
| zgc:92606 | 0.070 | rps12 | -0.062 |
| CR383676.1 | 0.070 | setb | -0.062 |
| nptna | 0.069 | snrpe | -0.061 |
| sv2a | 0.068 | snrpd1 | -0.061 |
| sh3gl2a | 0.067 | abcf1 | -0.060 |
| stmn4 | 0.067 | cirbpb | -0.059 |
| syn2a | 0.065 | hnrnpa1b | -0.058 |
| atp2b3b | 0.065 | syncrip | -0.058 |
| trim13 | 0.065 | snrpd2 | -0.058 |
| ppdpfb | 0.065 | snrpb | -0.058 |
| bada | 0.065 | snrpg | -0.058 |
| atp6v0cb | 0.064 | h2afvb | -0.058 |
| si:ch73-119p20.1 | 0.064 | ddx39ab | -0.057 |
| ier2b | 0.063 | dkc1 | -0.057 |
| pgk1 | 0.063 | nono | -0.057 |
| ndufa4 | 0.063 | hnrnph1l | -0.057 |
| gpia | 0.063 | npm1a | -0.056 |
| sparc | 0.063 | cbx3a | -0.056 |
| tob1a | 0.063 | hnrnpub | -0.055 |
| atp2b3a | 0.063 | rcc2 | -0.055 |
| pkma | 0.063 | hdac1 | -0.054 |
| syngr1a | 0.062 | si:dkey-56m19.5 | -0.054 |