MAX dimerization protein 3
ZFIN 
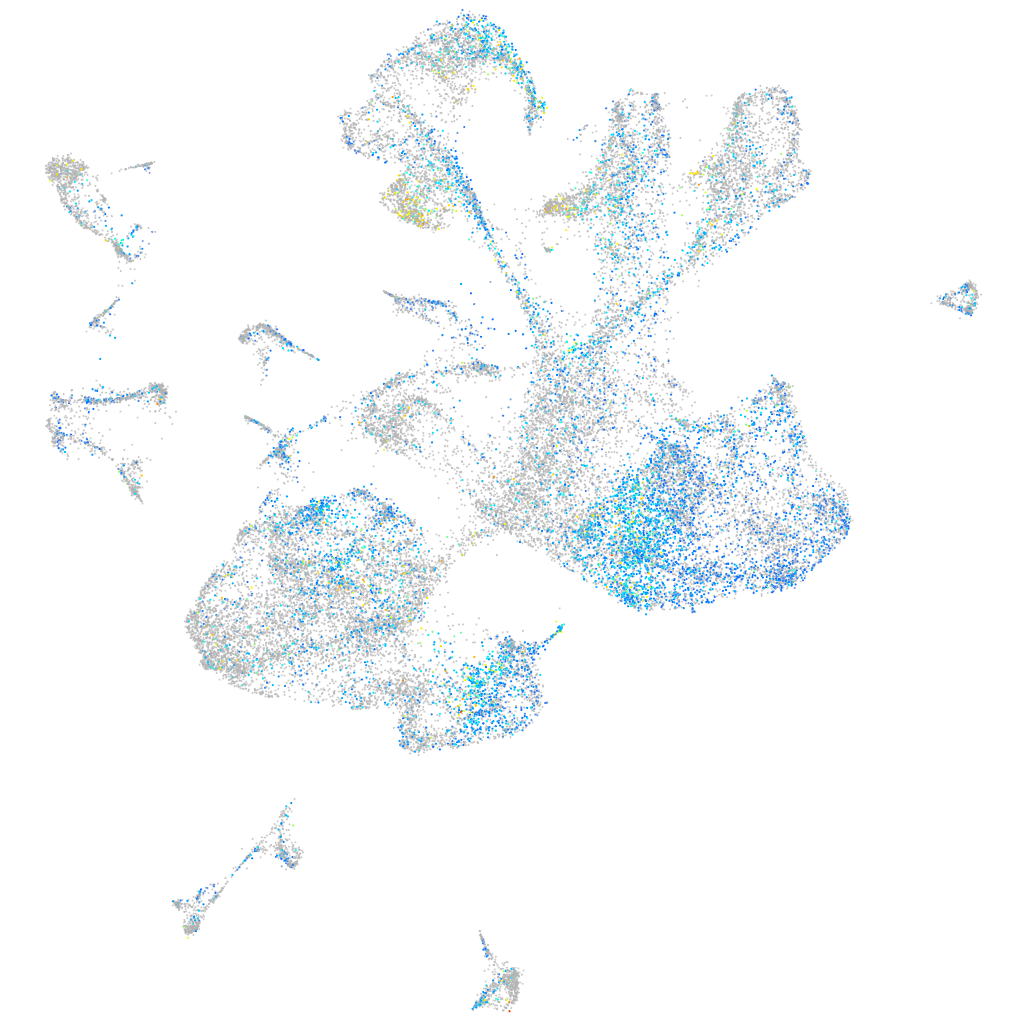
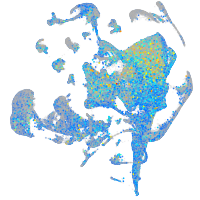
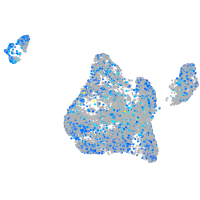
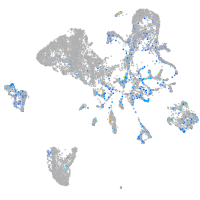
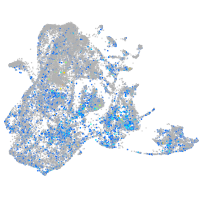
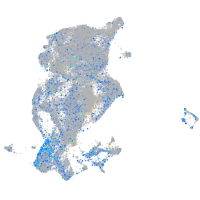
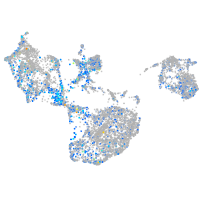
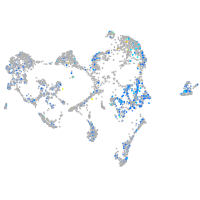
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| zgc:110540 | 0.326 | slc1a2b | -0.104 |
| mibp | 0.319 | gapdhs | -0.089 |
| rrm2 | 0.311 | ckbb | -0.085 |
| CABZ01005379.1 | 0.292 | cx43 | -0.085 |
| rrm1 | 0.275 | ptn | -0.085 |
| chaf1a | 0.268 | efhd1 | -0.084 |
| esco2 | 0.264 | slc6a1b | -0.084 |
| fbxo5 | 0.259 | acbd7 | -0.083 |
| dek | 0.258 | slc4a4a | -0.083 |
| pcna | 0.255 | ndrg3a | -0.082 |
| zgc:110216 | 0.253 | qki2 | -0.082 |
| zgc:165555.3 | 0.250 | mdkb | -0.081 |
| dut | 0.247 | cd63 | -0.081 |
| asf1ba | 0.247 | glula | -0.080 |
| si:dkey-261m9.17 | 0.246 | aldocb | -0.079 |
| slbp | 0.246 | hepacama | -0.079 |
| tk1 | 0.244 | CU467822.1 | -0.077 |
| stmn1a | 0.244 | gpr37l1b | -0.075 |
| si:dkey-108k21.10 | 0.239 | pvalb1 | -0.074 |
| ccne2 | 0.232 | anxa13 | -0.074 |
| cks1b | 0.231 | atp1a1b | -0.073 |
| lbr | 0.231 | si:ch211-66e2.5 | -0.073 |
| lig1 | 0.230 | pvalb2 | -0.072 |
| si:ch211-113a14.18 | 0.228 | itm2ba | -0.072 |
| ccna2 | 0.224 | fabp7a | -0.072 |
| suv39h1b | 0.220 | slc3a2a | -0.069 |
| LOC100330864 | 0.219 | smox | -0.069 |
| h3f3a | 0.218 | cdo1 | -0.069 |
| zgc:153405 | 0.218 | tob1b | -0.069 |
| mis12 | 0.218 | ptgdsb.2 | -0.068 |
| e2f8 | 0.217 | ppap2d | -0.068 |
| atad2 | 0.217 | IGLON5 | -0.066 |
| smc2 | 0.216 | mgll | -0.065 |
| nasp | 0.212 | gpm6bb | -0.065 |
| prim2 | 0.211 | rpz5 | -0.064 |