myotubularin related protein 3
ZFIN 
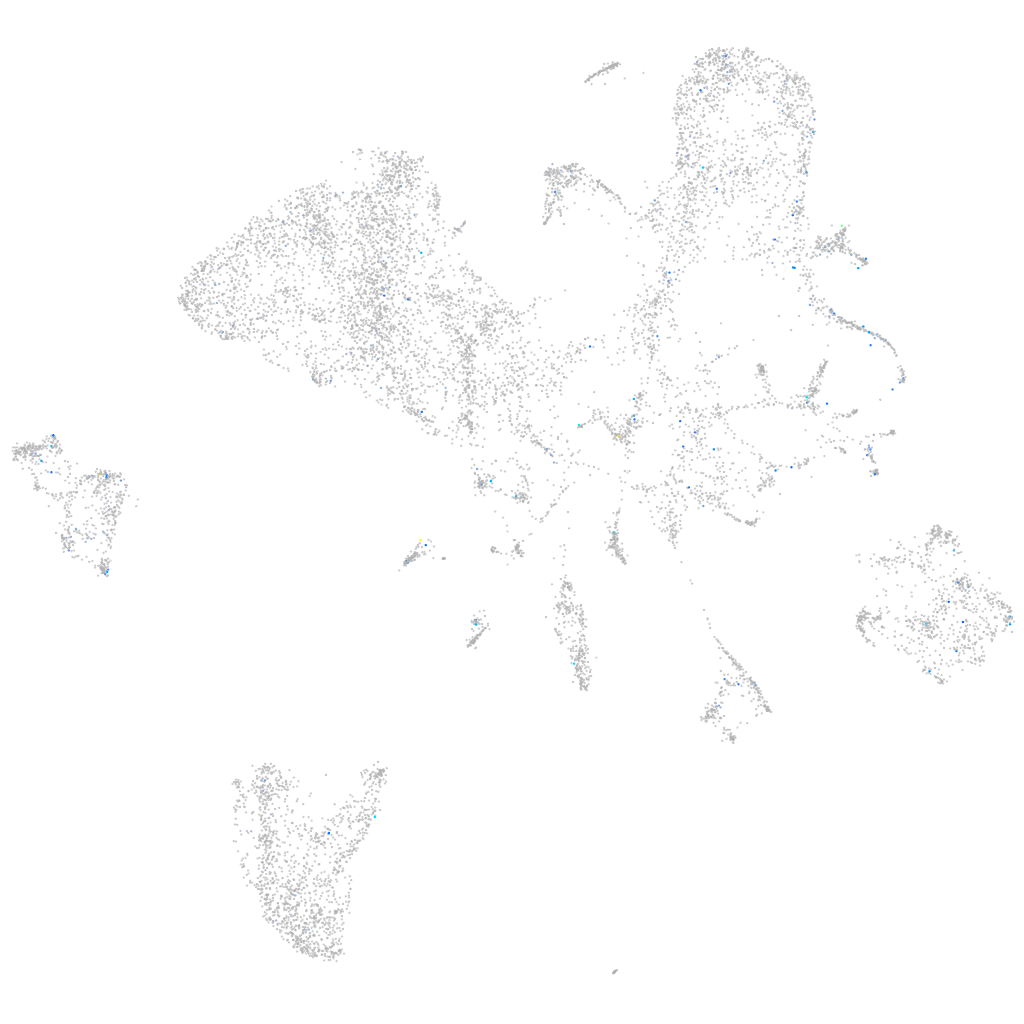
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cops6 | 0.180 | gamt | -0.065 |
| LOC103909114 | 0.164 | bhmt | -0.060 |
| XLOC-006316 | 0.153 | mat1a | -0.059 |
| LOC101884382 | 0.144 | aqp12 | -0.057 |
| klhl6 | 0.135 | gatm | -0.056 |
| LOC110439397 | 0.134 | grhprb | -0.055 |
| LOC101884949 | 0.132 | apoa1b | -0.054 |
| itga2.2 | 0.127 | kng1 | -0.054 |
| CABZ01078606.1 | 0.123 | agxtb | -0.051 |
| CABZ01079480.1 | 0.123 | ttc36 | -0.051 |
| CR749168.4 | 0.120 | ahcy | -0.050 |
| ncs1b | 0.118 | gpd1b | -0.049 |
| lhx8b | 0.117 | agxta | -0.049 |
| GABRR1 | 0.110 | rbp2b | -0.049 |
| BX842692.1 | 0.109 | hao1 | -0.049 |
| st18 | 0.109 | apom | -0.048 |
| ddr2b | 0.108 | fabp10a | -0.048 |
| LOC100538121 | 0.108 | serpina1 | -0.047 |
| b3gnt3.1 | 0.107 | apoa2 | -0.047 |
| zgc:111976 | 0.107 | serpinf2b | -0.047 |
| XLOC-033065 | 0.104 | tfa | -0.047 |
| BX001030.2 | 0.104 | pygl | -0.047 |
| LOC101886077 | 0.103 | LOC110437731 | -0.047 |
| gprc5bb | 0.101 | uox | -0.047 |
| BX649434.2 | 0.100 | zgc:123103 | -0.047 |
| LOC110439611 | 0.100 | serpina1l | -0.047 |
| pimr201 | 0.098 | cfhl4 | -0.046 |
| LOC110440119 | 0.097 | fgg | -0.046 |
| FP015862.1 | 0.097 | nipsnap3a | -0.046 |
| LOC101883531 | 0.096 | ambp | -0.046 |
| LOC101883046 | 0.096 | f2 | -0.046 |
| AL929396.1 | 0.096 | ttr | -0.046 |
| nlrc8 | 0.095 | fetub | -0.046 |
| CU550702.1 | 0.095 | rbp4 | -0.046 |
| si:ch211-37e10.1 | 0.095 | gc | -0.046 |