myotubularin related protein 14
ZFIN 
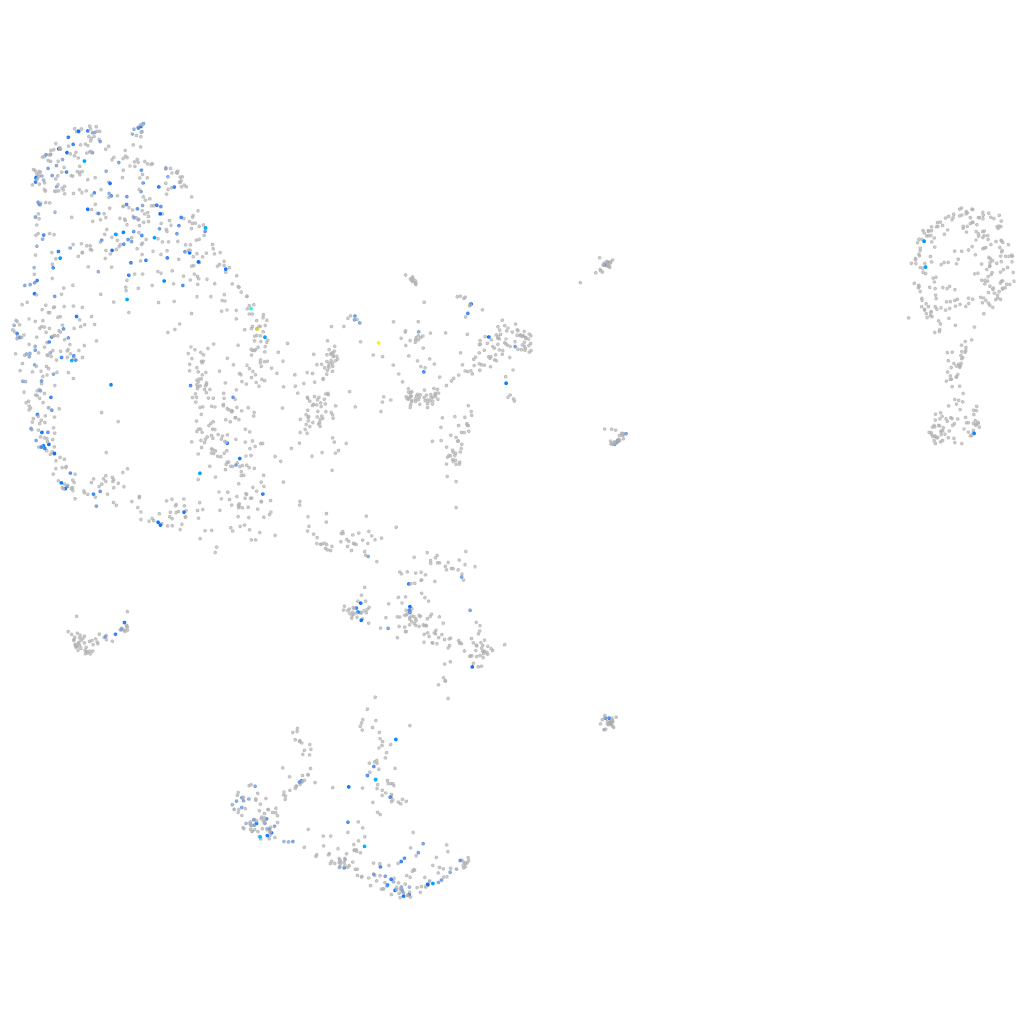
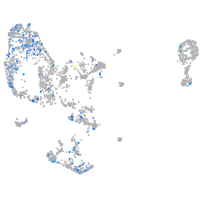
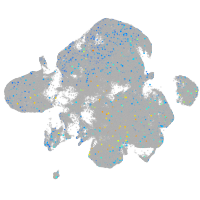
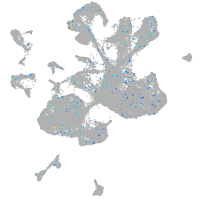
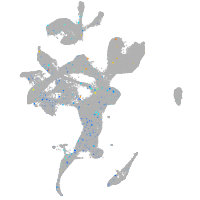
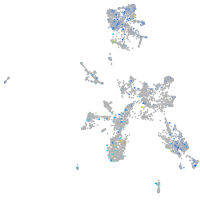
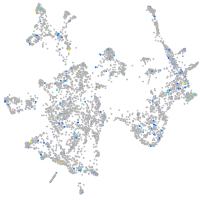
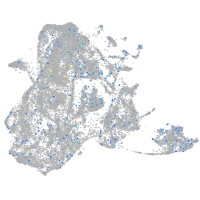
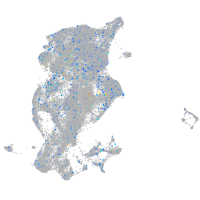
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| LOC101884106 | 0.224 | si:ch73-1a9.3 | -0.158 |
| ugt5c2 | 0.217 | ptmab | -0.157 |
| slc27a1b | 0.207 | si:ch211-222l21.1 | -0.150 |
| sdr16c5b | 0.205 | hmgn6 | -0.141 |
| glud1b | 0.203 | si:ch73-281n10.2 | -0.133 |
| mt-nd2 | 0.198 | hmgb3a | -0.130 |
| eps8l3a | 0.198 | h2afvb | -0.129 |
| si:ch211-139a5.9 | 0.197 | marcksl1b | -0.128 |
| mt-nd4 | 0.193 | hnrnpaba | -0.126 |
| si:ch211-71m22.3 | 0.193 | hspb1 | -0.125 |
| sod1 | 0.186 | marcksb | -0.125 |
| cox6a2 | 0.185 | nucks1a | -0.122 |
| mt-cyb | 0.184 | h3f3d | -0.118 |
| mt-nd1 | 0.184 | hmga1a | -0.118 |
| cox8b | 0.184 | id3 | -0.117 |
| si:ch211-253b8.5 | 0.183 | cirbpb | -0.116 |
| mt-atp6 | 0.183 | hmgb2a | -0.112 |
| lrrc66 | 0.182 | cirbpa | -0.112 |
| dapp1 | 0.181 | wu:fb97g03 | -0.112 |
| mt-co1 | 0.181 | hmgn2 | -0.112 |
| cdh17 | 0.179 | smarce1 | -0.112 |
| zgc:85777 | 0.176 | seta | -0.111 |
| gapdh | 0.176 | hmgb2b | -0.110 |
| gstp1 | 0.175 | apoeb | -0.109 |
| LOC101882070 | 0.175 | hnrnpa0b | -0.108 |
| zgc:110339 | 0.174 | apoc1 | -0.108 |
| suclg1 | 0.174 | hmgn7 | -0.108 |
| si:dkey-16p21.8 | 0.174 | snrpd3l | -0.107 |
| gabrr3a | 0.174 | ilf2 | -0.107 |
| COX3 | 0.174 | acin1a | -0.106 |
| rgrb | 0.174 | snrnp70 | -0.105 |
| calhm2 | 0.174 | msna | -0.104 |
| aldh6a1 | 0.174 | zgc:110425 | -0.103 |
| asl | 0.173 | snrpb | -0.102 |
| ltk | 0.172 | ewsr1a | -0.101 |