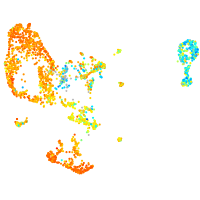"cytochrome b, mitochondrial"
ZFIN 
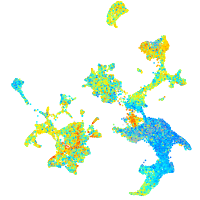
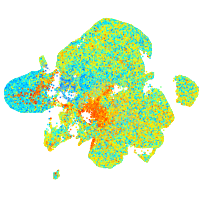
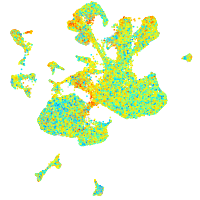
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| mt-co2 | 0.536 | cirbpb | -0.397 |
| COX3 | 0.525 | fam189a1 | -0.354 |
| mt-nd1 | 0.486 | abhd6b | -0.304 |
| zgc:86839 | 0.429 | nnr | -0.296 |
| mt-co1 | 0.409 | surf2 | -0.296 |
| taz | 0.407 | rps18 | -0.294 |
| mt-atp6 | 0.406 | fam160b2 | -0.294 |
| ifrd1 | 0.387 | nvl | -0.294 |
| znf384l | 0.386 | sesn3 | -0.291 |
| ier2a | 0.382 | CABZ01084225.1 | -0.286 |
| calm1a | 0.380 | rpl5b | -0.284 |
| naa40 | 0.378 | acin1a | -0.282 |
| strada | 0.377 | kcnc1b | -0.282 |
| flncb | 0.374 | zp3d.2 | -0.282 |
| NC-002333.17 | 0.373 | eif2s2 | -0.280 |
| mt-nd4 | 0.371 | hmgb1a | -0.276 |
| fuca1.1 | 0.370 | si:ch211-269k10.2 | -0.275 |
| smdt1b | 0.369 | pax8 | -0.275 |
| tmem147 | 0.362 | hmga1a | -0.273 |
| mt-nd2 | 0.360 | zrsr2 | -0.270 |
| ints11 | 0.360 | slu7 | -0.270 |
| btr04 | 0.359 | eif3s10 | -0.265 |
| atp6v0cb | 0.356 | CABZ01084081.1 | -0.265 |
| UBA6 | 0.356 | mns1 | -0.264 |
| snx9a | 0.355 | klhl14 | -0.264 |
| dpm2 | 0.352 | FP236331.1 | -0.263 |
| derl2 | 0.351 | mhc1zaa | -0.263 |
| npm2b | 0.351 | crygn1 | -0.263 |
| gnptg | 0.350 | CT573178.1 | -0.263 |
| fgf12b | 0.350 | trabd2b | -0.263 |
| cadpsb | 0.349 | bbc3 | -0.263 |
| dguok | 0.349 | BX293567.1 | -0.263 |
| ginm1 | 0.349 | BX323575.1 | -0.263 |
| pdss1 | 0.347 | BX681417.2 | -0.263 |
| selenom | 0.344 | CABZ01051877.1 | -0.263 |