muscle segment homeobox 2a
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
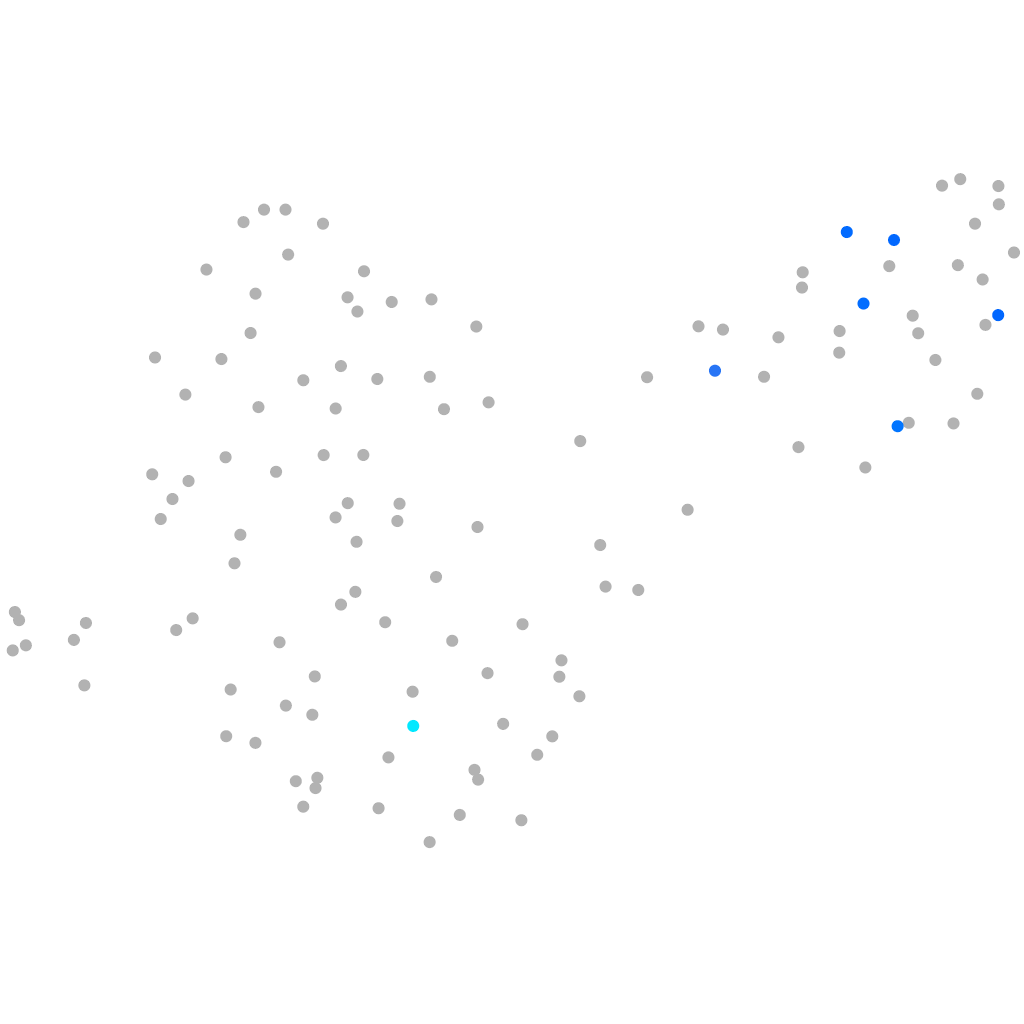
axial mesoderm |
muscle 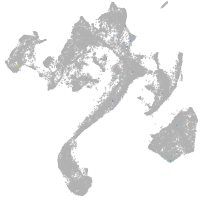 |
mural cells / non-skeletal muscle 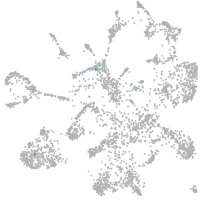 |
mesenchyme 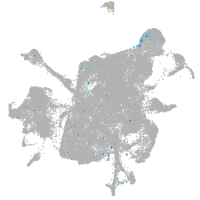 |
hematopoietic / vasculature 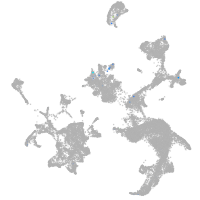 |
pronephros  |
neural |
spinal cord / glia |
eye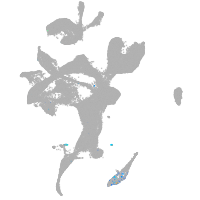 |
taste / olfactory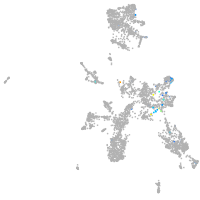 |
otic / lateral line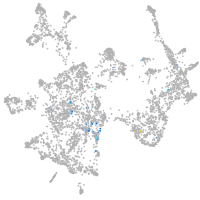 |
epidermis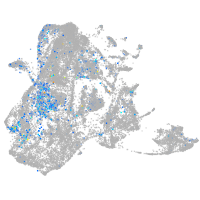 |
periderm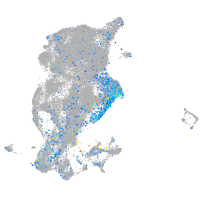 |
fin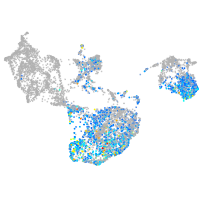 |
ionocytes / mucous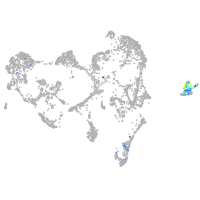 |
pigment cells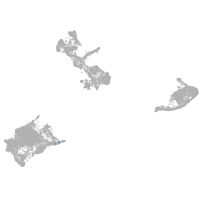 |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| wfdc2 | 0.809 | rps18 | -0.265 |
| spidr | 0.786 | nop56 | -0.264 |
| slc2a10 | 0.771 | ncl | -0.262 |
| tekt1 | 0.747 | tpx2 | -0.243 |
| gprc5c | 0.735 | nop58 | -0.241 |
| prokr1a | 0.729 | marcksl1b | -0.237 |
| adm2a | 0.729 | acin1b | -0.237 |
| ccdc39 | 0.716 | ebna1bp2 | -0.236 |
| lmo7a | 0.690 | baz1b | -0.235 |
| armc8 | 0.678 | cdk11b | -0.230 |
| slc7a2 | 0.670 | si:ch73-281n10.2 | -0.229 |
| zgc:114181 | 0.669 | zgc:56676 | -0.228 |
| sult2st3 | 0.669 | snrpa | -0.223 |
| adamts15a | 0.669 | mphosph10 | -0.219 |
| BX323824.1 | 0.669 | fth1a | -0.218 |
| BX539336.1 | 0.669 | NC-002333.4 | -0.217 |
| jph3 | 0.669 | npm1a | -0.216 |
| si:ch211-66e2.5 | 0.669 | mettl5 | -0.214 |
| si:dkeyp-92c9.4 | 0.669 | mki67 | -0.213 |
| slc22a5 | 0.669 | ddx18 | -0.211 |
| slc7a8b | 0.669 | prkrip1 | -0.209 |
| steap4 | 0.669 | luc7l3 | -0.208 |
| thbs4b | 0.669 | arf1 | -0.207 |
| tlr18 | 0.669 | htatsf1 | -0.206 |
| tmem72 | 0.669 | ppig | -0.206 |
| angptl1a | 0.669 | rrp1 | -0.205 |
| si:dkeyp-69c1.9 | 0.669 | cactin | -0.205 |
| slc13a1 | 0.669 | acin1a | -0.205 |
| muc5.3 | 0.669 | serbp1b | -0.205 |
| six6a | 0.669 | fbl | -0.202 |
| tmtc2b | 0.669 | srrm1 | -0.201 |
| birc6-as2 | 0.669 | pttg1 | -0.200 |
| BX511120.1 | 0.669 | rpa3 | -0.198 |
| cfap126 | 0.669 | thoc7 | -0.197 |
| cfap52 | 0.669 | gar1 | -0.196 |