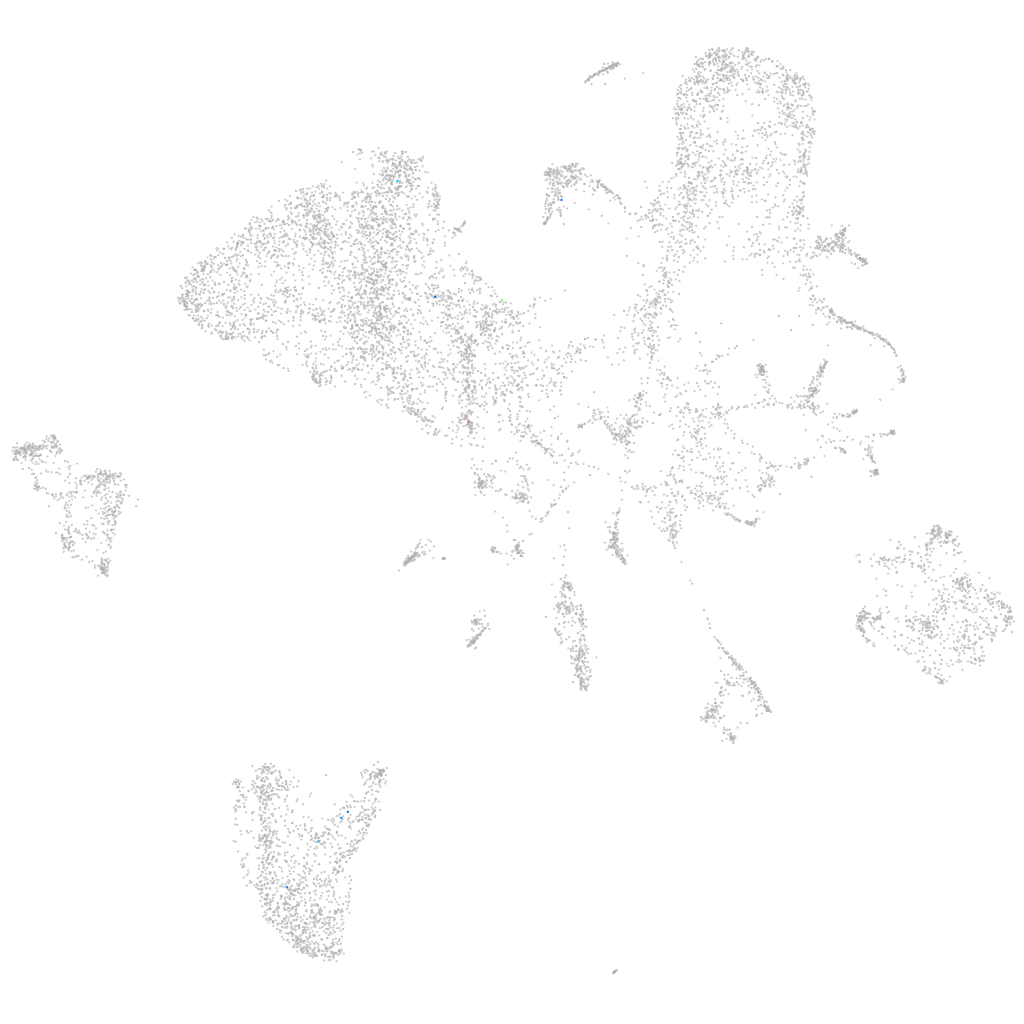
moesin b
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros 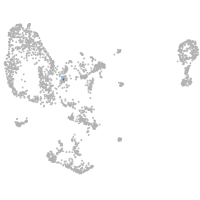 |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line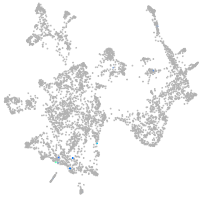 |
epidermis |
periderm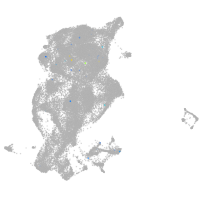 |
fin |
ionocytes / mucous |
pigment cells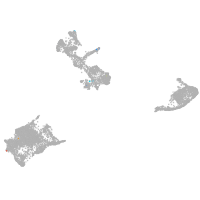 |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cilp2 | 0.627 | rpl13a | -0.052 |
| inpp5kb | 0.525 | rpl12 | -0.051 |
| trpm1a | 0.516 | rps15a | -0.047 |
| sycp1 | 0.498 | hsp90ab1 | -0.046 |
| pmelb | 0.493 | rpl26 | -0.046 |
| tmem136b | 0.488 | rpl10 | -0.044 |
| CT573799.2 | 0.473 | rps21 | -0.040 |
| gpr176 | 0.453 | rpl30 | -0.039 |
| dchs1a | 0.440 | rps28 | -0.037 |
| mitfa | 0.439 | naca | -0.036 |
| pcolceb | 0.425 | rack1 | -0.036 |
| LOC101884756 | 0.400 | ybx1 | -0.034 |
| tm4sf18 | 0.389 | rpl34 | -0.033 |
| triobpa | 0.372 | slc25a3b | -0.031 |
| BX927308.2 | 0.344 | rpl14 | -0.030 |
| CU681836.1 | 0.340 | rps27.2 | -0.029 |
| scpp5 | 0.329 | eif3g | -0.027 |
| mxra5b | 0.327 | pabpc1a | -0.027 |
| proca1 | 0.316 | ptmab | -0.027 |
| ppfia2 | 0.316 | romo1 | -0.027 |
| crybb1l2 | 0.279 | eif1b | -0.026 |
| si:dkey-262g12.14 | 0.277 | ppiaa | -0.026 |
| col4a3 | 0.267 | eif3m | -0.026 |
| si:dkey-35i13.1 | 0.265 | rpl23a | -0.026 |
| cfp | 0.263 | oaz1a | -0.026 |
| si:dkey-21e2.15 | 0.263 | cox6c | -0.025 |
| CU651662.1 | 0.260 | tma7 | -0.025 |
| rgra | 0.259 | cox8a | -0.025 |
| si:dkey-112e17.1 | 0.258 | rpl29 | -0.025 |
| fgf12a | 0.253 | btf3 | -0.025 |
| trim63b | 0.251 | cirbpb | -0.025 |
| thbs1b | 0.250 | atp5mc3b | -0.025 |
| gabra2a | 0.238 | NC-002333.4 | -0.025 |
| col15a1b | 0.237 | ndufb8 | -0.025 |
| ppp1r9a | 0.233 | myl12.1 | -0.024 |