mesothelin a
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros 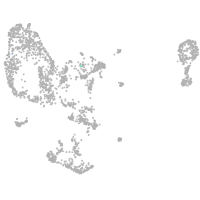 |
neural |
spinal cord / glia |
eye |
taste / olfactory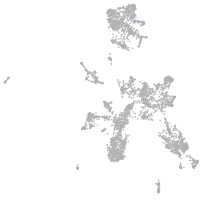 |
otic / lateral line |
epidermis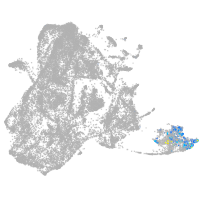 |
periderm |
fin |
ionocytes / mucous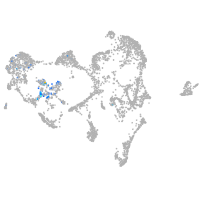 |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| sim2.1 | 0.990 | cox4i1 | -0.062 |
| angptl1b | 0.959 | rpl5b | -0.057 |
| slc5a8 | 0.906 | atp5f1e | -0.053 |
| tmprss13a | 0.846 | eif3f | -0.053 |
| fbln5 | 0.817 | atp5fa1 | -0.046 |
| LOC110439013 | 0.790 | rps26l | -0.043 |
| agrp | 0.786 | atp5if1a | -0.040 |
| pi15a | 0.689 | cycsb | -0.040 |
| si:ch211-244b2.4 | 0.677 | cirbpa | -0.039 |
| loxa | 0.658 | tma7 | -0.039 |
| ptchd4 | 0.623 | uqcrfs1 | -0.039 |
| si:ch211-136a13.1 | 0.623 | rpl29 | -0.038 |
| AL929048.1 | 0.615 | hsp90ab1 | -0.038 |
| zgc:153157 | 0.610 | calm2b | -0.037 |
| glis1b | 0.595 | hmgn7 | -0.036 |
| c1qtnf12 | 0.589 | hmgn2 | -0.036 |
| ecm2 | 0.589 | mt-nd2 | -0.036 |
| card14 | 0.582 | snrpf | -0.035 |
| CABZ01039096.1 | 0.565 | ywhaba | -0.035 |
| crispld1a | 0.549 | edf1 | -0.035 |
| thbs3a | 0.545 | srsf5a | -0.035 |
| CABZ01092282.1 | 0.529 | ndufb9 | -0.035 |
| adgrd1 | 0.520 | mt-nd4 | -0.034 |
| FP102018.1 | 0.514 | cyc1 | -0.034 |
| aspn | 0.511 | eif2s2 | -0.034 |
| si:ch211-43f4.1 | 0.509 | romo1 | -0.034 |
| unm-sa808 | 0.504 | eif3ba | -0.033 |
| col27a1b | 0.489 | eif3g | -0.033 |
| spon2b | 0.479 | dynll1 | -0.033 |
| plekhg2 | 0.474 | ndufab1a | -0.033 |
| si:dkey-12l12.1 | 0.466 | ube2v2 | -0.033 |
| LOC100535170 | 0.464 | dad1 | -0.033 |
| arap1a | 0.457 | cnbpb | -0.033 |
| spon2a | 0.450 | mt-nd5 | -0.032 |
| sema4ga | 0.450 | ppp2cb | -0.032 |