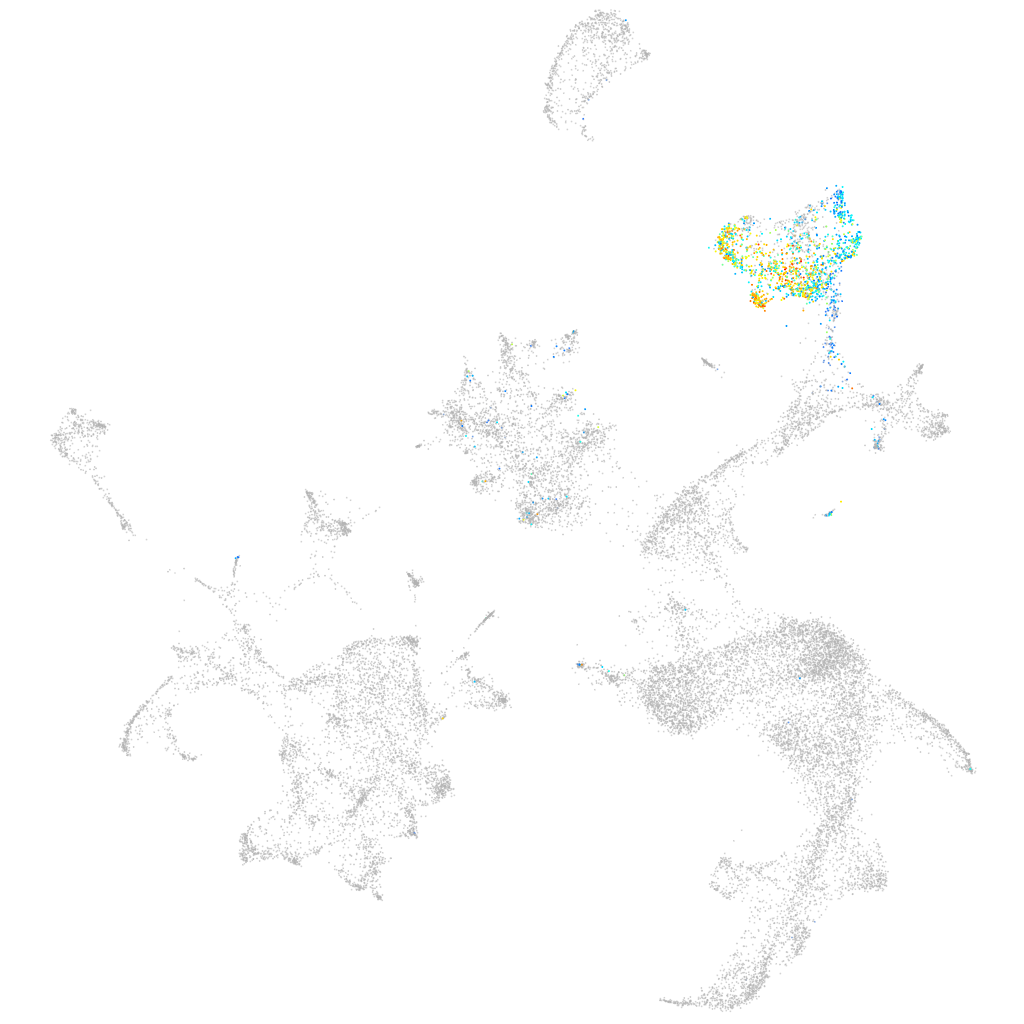
"macrophage expressed 1, tandem duplicate 1"
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| havcr1 | 0.794 | hbae1.1 | -0.240 |
| cmklr1 | 0.777 | hbae1.3 | -0.229 |
| si:dkey-5n18.1 | 0.767 | hbae3 | -0.227 |
| ctss2.2 | 0.746 | hbbe1.3 | -0.227 |
| mfap4 | 0.731 | hbbe2 | -0.227 |
| lgals3bpb | 0.727 | hbbe1.1 | -0.217 |
| cx32.2 | 0.717 | cahz | -0.209 |
| lgals2a | 0.701 | fth1a | -0.207 |
| si:zfos-1069f5.1 | 0.697 | zgc:56095 | -0.199 |
| tnfb | 0.672 | hbbe1.2 | -0.198 |
| ccl34b.1 | 0.671 | hemgn | -0.196 |
| slc7a7 | 0.669 | lmo2 | -0.193 |
| ctsl.1 | 0.661 | alas2 | -0.184 |
| CU499330.1 | 0.658 | si:ch211-250g4.3 | -0.182 |
| si:ch211-147m6.1 | 0.642 | slc4a1a | -0.173 |
| slc3a2a | 0.641 | nt5c2l1 | -0.169 |
| tspan36 | 0.639 | creg1 | -0.166 |
| lygl1 | 0.638 | blvrb | -0.161 |
| marco | 0.636 | zgc:163057 | -0.160 |
| tnfsf12 | 0.628 | epb41b | -0.160 |
| si:ch211-212k18.7 | 0.626 | si:ch211-207c6.2 | -0.155 |
| ms4a17a.10 | 0.624 | tspo | -0.152 |
| ctsc | 0.619 | nmt1b | -0.149 |
| ctsba | 0.618 | aqp1a.1 | -0.149 |
| gm2a | 0.617 | tmod4 | -0.142 |
| c1qc | 0.615 | plac8l1 | -0.141 |
| itgae.1 | 0.614 | arl4aa | -0.133 |
| rnaset2l | 0.614 | histh1l | -0.133 |
| si:ch211-194m7.3 | 0.614 | znfl2a | -0.132 |
| nfkbiaa | 0.595 | sptb | -0.132 |
| ccl34a.4 | 0.594 | hbae5 | -0.131 |
| si:ch73-158p21.3 | 0.593 | uros | -0.129 |
| grna | 0.590 | si:ch211-227m13.1 | -0.129 |
| cxcr3.2 | 0.590 | tfr1a | -0.128 |
| c1qb | 0.583 | si:ch211-222l21.1 | -0.126 |