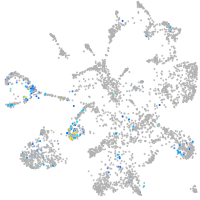
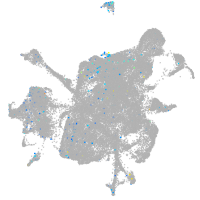
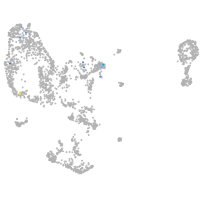
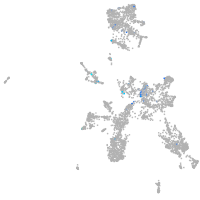
matrix metallopeptidase 17a
ZFIN 
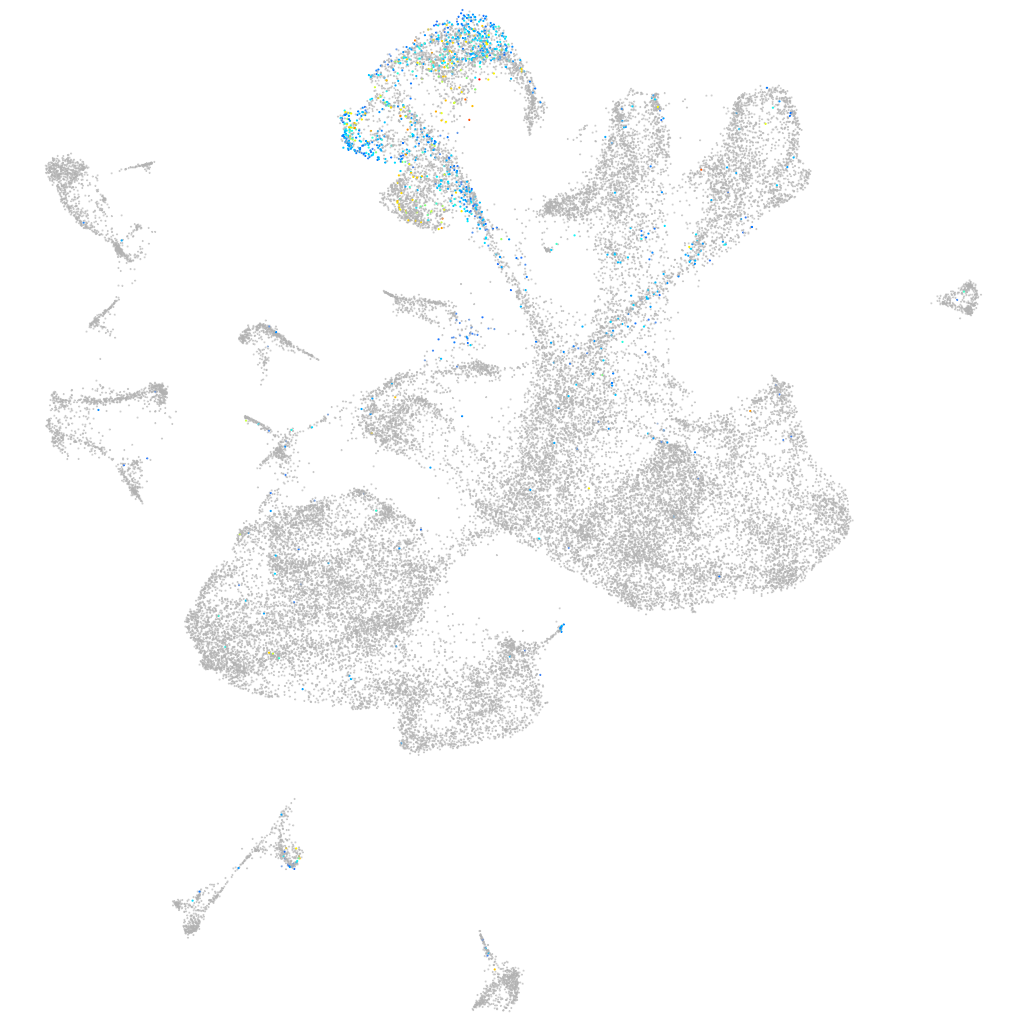
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| slc18a3a | 0.305 | XLOC-003690 | -0.158 |
| chodl | 0.284 | id1 | -0.150 |
| tbx3a | 0.270 | mdka | -0.140 |
| slit3 | 0.266 | her15.1 | -0.136 |
| lhx4 | 0.248 | cldn5a | -0.129 |
| sv2c | 0.248 | sox3 | -0.128 |
| tbx20 | 0.245 | notch3 | -0.119 |
| isl1 | 0.245 | XLOC-003689 | -0.118 |
| slc5a7a | 0.243 | vamp3 | -0.116 |
| prph | 0.242 | hmgb2a | -0.114 |
| map1b | 0.241 | btg2 | -0.110 |
| isl2a | 0.234 | sdc4 | -0.110 |
| CABZ01074363.1 | 0.227 | msi1 | -0.108 |
| ache | 0.224 | tspan7 | -0.108 |
| si:ch211-247j9.1 | 0.218 | sparc | -0.107 |
| chata | 0.216 | sox19a | -0.105 |
| SLC35D3 | 0.216 | COX7A2 (1 of many) | -0.104 |
| tac1 | 0.215 | her4.1 | -0.104 |
| lmo4b | 0.215 | her4.2 | -0.103 |
| nefmb | 0.213 | si:ch73-281n10.2 | -0.102 |
| agrn | 0.212 | her12 | -0.101 |
| onecut1 | 0.210 | XLOC-003692 | -0.100 |
| cntn2 | 0.208 | msna | -0.099 |
| chga | 0.208 | sb:cb81 | -0.098 |
| islr2 | 0.205 | cd82a | -0.097 |
| gap43 | 0.200 | si:dkey-151g10.6 | -0.097 |
| aplp1 | 0.198 | sox2 | -0.097 |
| elavl4 | 0.196 | si:ch211-286b5.5 | -0.094 |
| rtn1b | 0.194 | gpm6bb | -0.091 |
| gng3 | 0.192 | gfap | -0.091 |
| stmn2a | 0.190 | zgc:56493 | -0.091 |
| mllt11 | 0.187 | fosab | -0.089 |
| inab | 0.185 | rps20 | -0.089 |
| stmn4l | 0.185 | pgrmc1 | -0.088 |
| nrg1 | 0.184 | rplp1 | -0.087 |