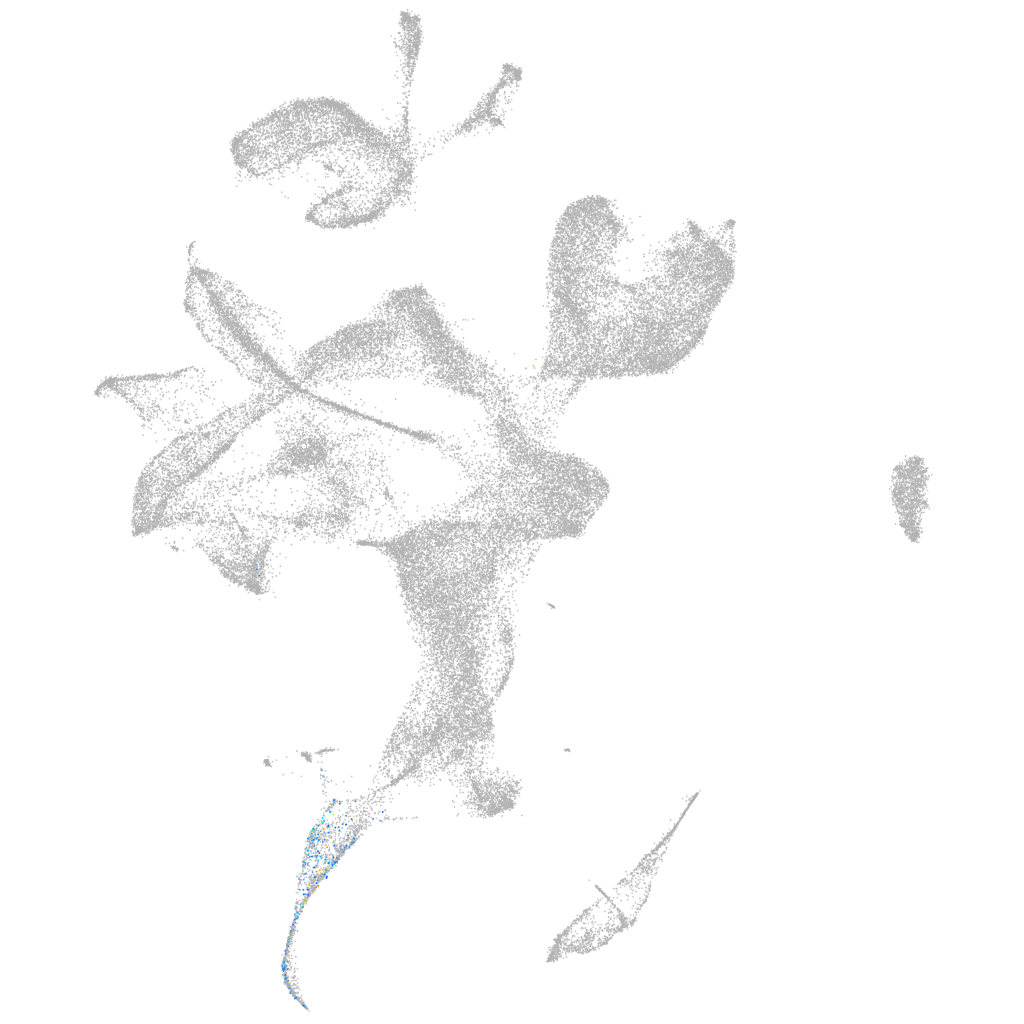
melan-A
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| tyrp1b | 0.326 | ptmab | -0.119 |
| pmelb | 0.319 | si:ch73-1a9.3 | -0.113 |
| dct | 0.318 | ptmaa | -0.076 |
| slc45a2 | 0.304 | tuba1c | -0.071 |
| oca2 | 0.304 | hmgn6 | -0.070 |
| pmela | 0.303 | marcksl1b | -0.069 |
| rab38 | 0.293 | si:ch211-137a8.4 | -0.064 |
| tyrp1a | 0.290 | gpm6aa | -0.063 |
| tspan36 | 0.290 | hmgn2 | -0.061 |
| gpr143 | 0.287 | hnrnpaba | -0.060 |
| fabp11b | 0.284 | h2afva | -0.059 |
| slc24a5 | 0.269 | hmgb1a | -0.059 |
| tspan10 | 0.269 | fabp7a | -0.056 |
| tyr | 0.264 | sumo3a | -0.054 |
| cracr2ab | 0.263 | hmgb1b | -0.054 |
| zgc:110591 | 0.262 | si:ch1073-429i10.3.1 | -0.054 |
| rgrb | 0.252 | cirbpb | -0.052 |
| agtrap | 0.249 | nova2 | -0.052 |
| bace2 | 0.243 | hmgb3a | -0.052 |
| fgfbp1a | 0.242 | tuba1a | -0.051 |
| rab27a | 0.238 | hmga1a | -0.050 |
| mb | 0.237 | si:ch73-281n10.2 | -0.050 |
| tm6sf2 | 0.235 | si:ch211-222l21.1 | -0.049 |
| msnb | 0.235 | si:ch211-288g17.3 | -0.049 |
| pttg1ipb | 0.234 | marcksb | -0.049 |
| triobpa | 0.234 | epb41a | -0.048 |
| gstp1 | 0.234 | anp32e | -0.048 |
| col4a5 | 0.233 | marcksl1a | -0.047 |
| qdpra | 0.232 | hnrnpabb | -0.046 |
| mitfa | 0.232 | gapdhs | -0.045 |
| si:ch73-86n18.1 | 0.228 | cadm3 | -0.045 |
| FP085398.1 | 0.223 | h3f3d | -0.045 |
| sytl2b | 0.223 | mdkb | -0.044 |
| si:dkey-21a6.5 | 0.220 | hdac1 | -0.044 |
| col4a6 | 0.220 | rorb | -0.044 |