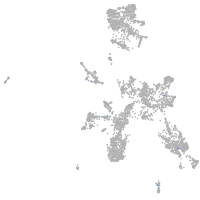
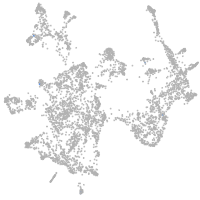
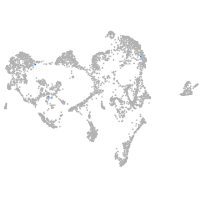
melanocyte inducing transcription factor a
ZFIN 
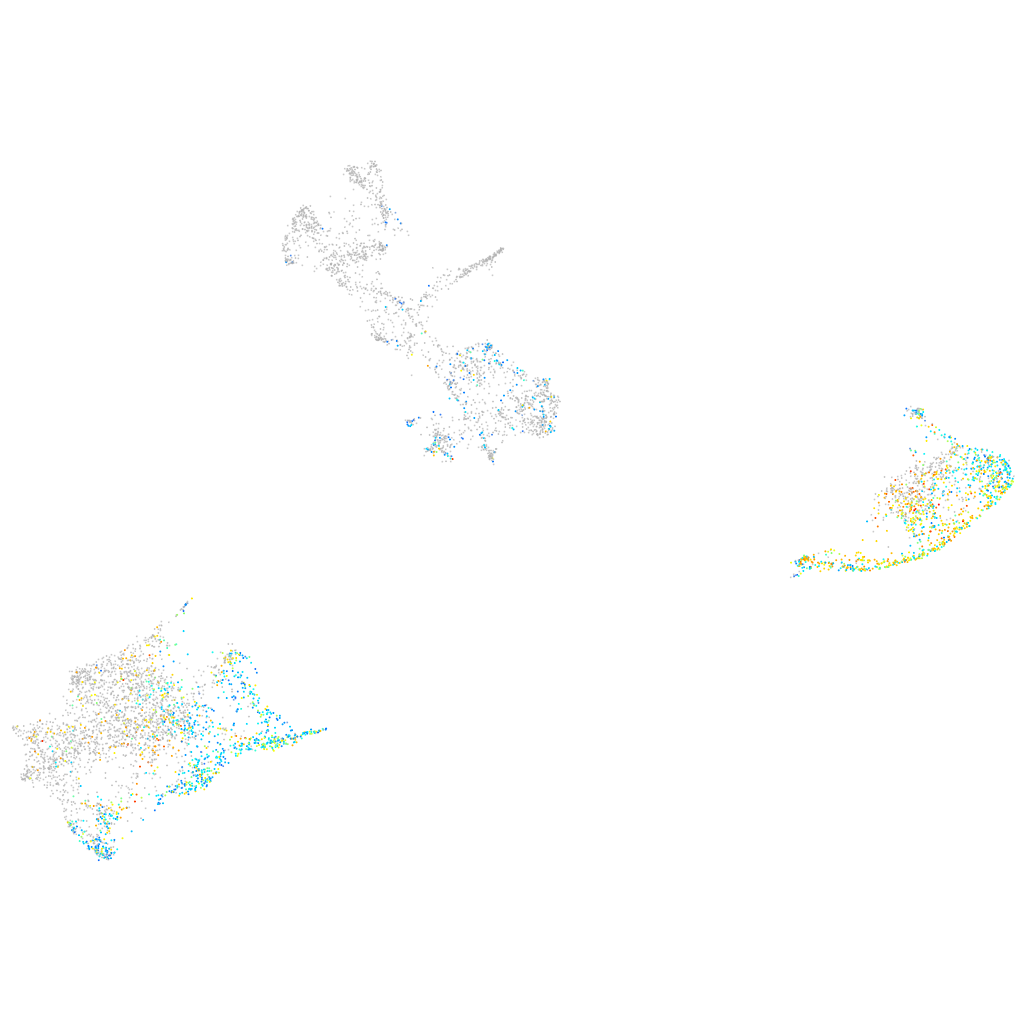
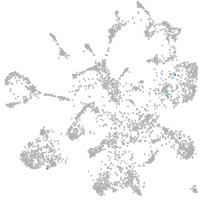
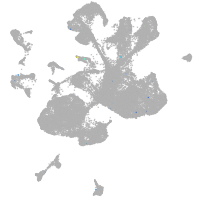
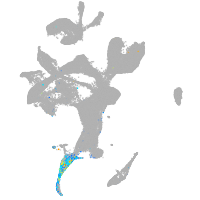
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| tfap2e | 0.439 | defbl1 | -0.292 |
| tspan36 | 0.424 | apoda.1 | -0.288 |
| tyr | 0.420 | si:ch211-256m1.8 | -0.264 |
| tmem243b | 0.418 | gpnmb | -0.264 |
| pmela | 0.414 | lypc | -0.255 |
| dct | 0.408 | si:dkey-197i20.6 | -0.251 |
| tyrp1b | 0.403 | mdkb | -0.243 |
| qdpra | 0.400 | unm-sa821 | -0.239 |
| rabl6b | 0.398 | alx4a | -0.232 |
| tyrp1a | 0.397 | si:ch73-89b15.3 | -0.228 |
| si:ch73-389b16.1 | 0.394 | akap12a | -0.225 |
| zgc:91968 | 0.383 | s100v2 | -0.219 |
| tspan10 | 0.381 | alx4b | -0.215 |
| msx1b | 0.381 | cdh11 | -0.211 |
| SPAG9 | 0.375 | ponzr1 | -0.208 |
| oca2 | 0.371 | sytl2b | -0.205 |
| lamp1a | 0.368 | slc25a36a | -0.203 |
| slc3a2a | 0.368 | ptmaa | -0.201 |
| pah | 0.366 | guk1a | -0.200 |
| kita | 0.364 | prx | -0.199 |
| slc24a5 | 0.362 | alx1 | -0.199 |
| slc37a2 | 0.362 | tcirg1a | -0.194 |
| tmem243a | 0.357 | cx43 | -0.191 |
| pcbd1 | 0.357 | ltk | -0.179 |
| rab38 | 0.352 | myof | -0.179 |
| gstt1a | 0.351 | sparc | -0.178 |
| slc45a2 | 0.350 | pltp | -0.178 |
| C12orf75 | 0.337 | fhl2a | -0.177 |
| syngr1a | 0.335 | sdc2 | -0.174 |
| bace2 | 0.333 | fhl3a | -0.172 |
| sypl2b | 0.325 | CT737162.3 | -0.172 |
| pttg1ipb | 0.323 | tmem179b | -0.172 |
| cdh1 | 0.320 | CABZ01072077.1 | -0.171 |
| mtbl | 0.320 | sulf2b | -0.169 |
| slc7a5 | 0.320 | tpd52l1 | -0.169 |