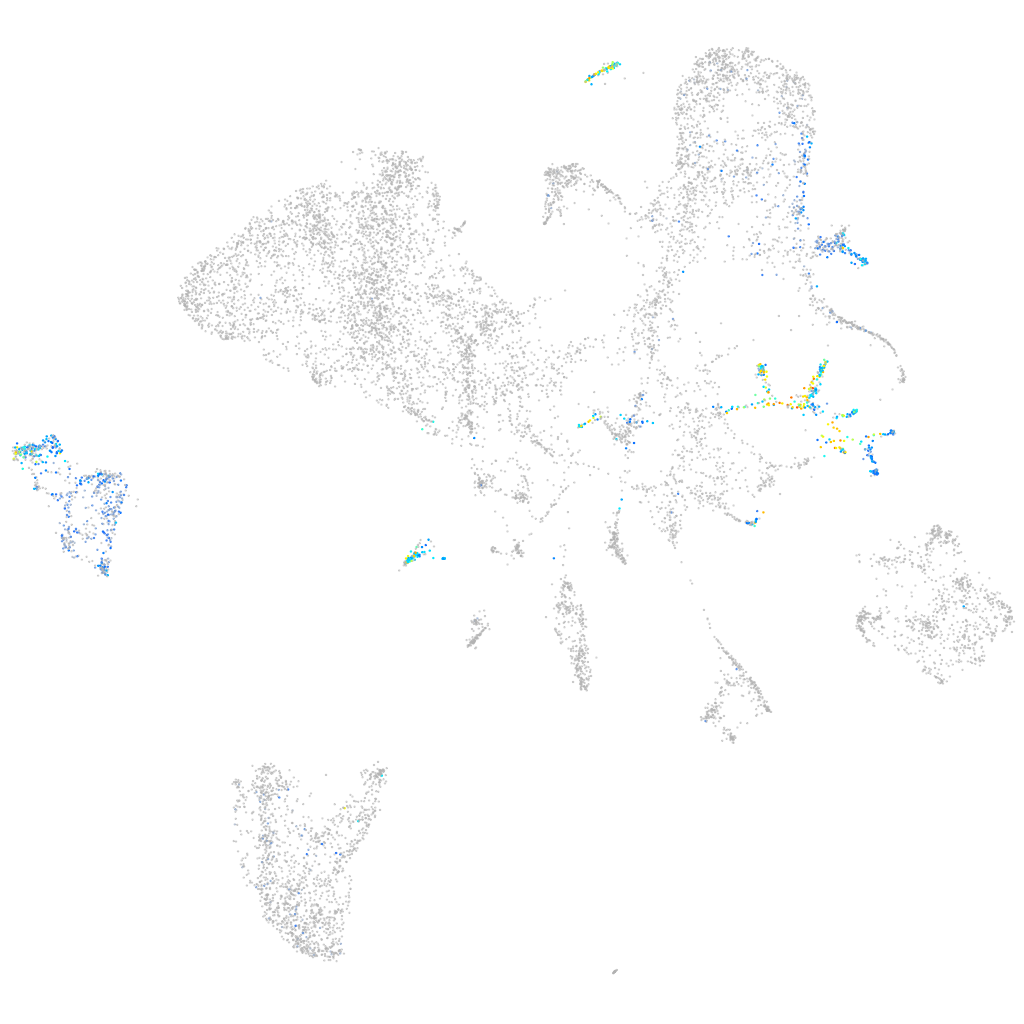
microRNA 375-2
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia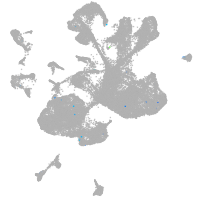 |
eye |
taste / olfactory |
otic / lateral line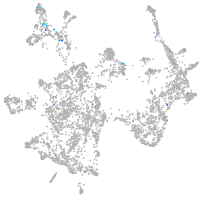 |
epidermis |
periderm |
fin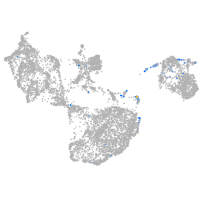 |
ionocytes / mucous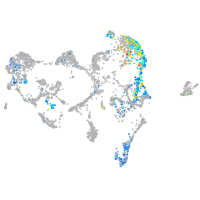 |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| scg3 | 0.577 | gamt | -0.259 |
| fev | 0.573 | ahcy | -0.248 |
| neurod1 | 0.518 | gapdh | -0.247 |
| elovl1a | 0.509 | gatm | -0.229 |
| mir7a-1 | 0.471 | bhmt | -0.223 |
| calm1b | 0.466 | mat1a | -0.215 |
| pax6b | 0.461 | fbp1b | -0.210 |
| insm1b | 0.450 | apoa1b | -0.208 |
| hepacam2 | 0.442 | apoc2 | -0.207 |
| egr4 | 0.435 | hspe1 | -0.205 |
| c2cd4a | 0.423 | apoc1 | -0.204 |
| insm1a | 0.416 | si:dkey-16p21.8 | -0.204 |
| pcsk1nl | 0.413 | apoa4b.1 | -0.201 |
| scgn | 0.411 | afp4 | -0.200 |
| nkx2.2a | 0.395 | gstt1a | -0.196 |
| cldnh | 0.395 | aqp12 | -0.194 |
| zgc:101731 | 0.393 | cx32.3 | -0.194 |
| tmed4 | 0.390 | agxta | -0.192 |
| rprmb | 0.389 | agxtb | -0.189 |
| gapdhs | 0.388 | tfa | -0.185 |
| dll4 | 0.386 | glud1b | -0.185 |
| arxa | 0.375 | hspd1 | -0.184 |
| syt1a | 0.371 | wu:fj16a03 | -0.184 |
| pygma | 0.368 | adka | -0.183 |
| hpcal1 | 0.367 | apoa2 | -0.182 |
| id4 | 0.367 | aldh7a1 | -0.181 |
| vamp2 | 0.365 | gstr | -0.179 |
| si:dkey-153k10.9 | 0.361 | aldh6a1 | -0.179 |
| lysmd2 | 0.359 | gcshb | -0.179 |
| tmem59 | 0.359 | apobb.1 | -0.179 |
| rnasekb | 0.358 | scp2a | -0.178 |
| pax4 | 0.355 | serpina1 | -0.177 |
| capsla | 0.353 | ftcd | -0.177 |
| anxa4 | 0.352 | serpinf2b | -0.175 |
| ret | 0.351 | serpina1l | -0.174 |