microRNA 181b-3
ZFIN 
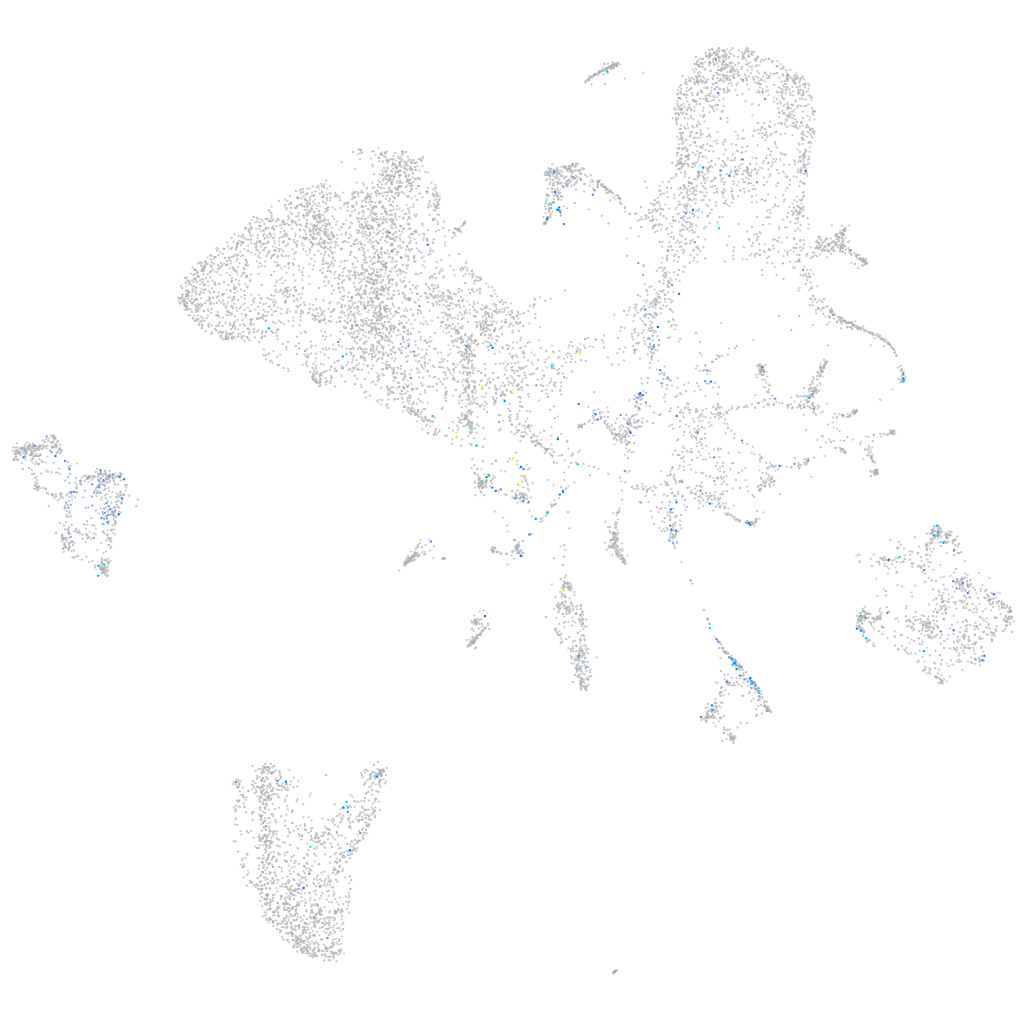
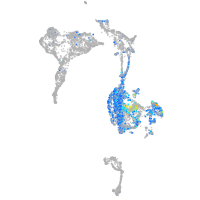
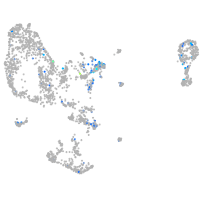
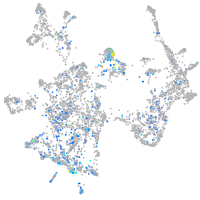
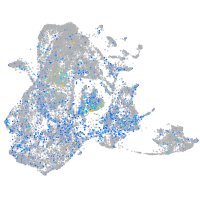
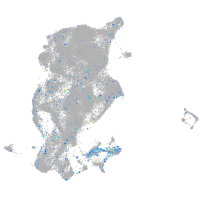
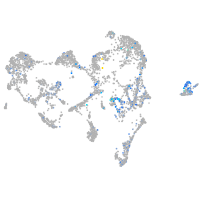
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| sox14 | 0.170 | gapdh | -0.110 |
| hapln3 | 0.153 | gamt | -0.099 |
| BX899181.1 | 0.149 | suclg1 | -0.095 |
| ptchd4 | 0.146 | apoa4b.1 | -0.094 |
| sox2 | 0.140 | gpx4a | -0.094 |
| CEP170B (1 of many) | 0.140 | scp2a | -0.094 |
| sox12 | 0.140 | fbp1b | -0.090 |
| gabrz | 0.138 | gatm | -0.090 |
| gfra2b | 0.135 | dhrs9 | -0.088 |
| XLOC-003052 | 0.135 | aldob | -0.088 |
| lama5 | 0.133 | pklr | -0.087 |
| CR749763.3 | 0.132 | glud1b | -0.087 |
| XLOC-022188 | 0.131 | mat1a | -0.086 |
| ptprz1a | 0.131 | dap | -0.086 |
| si:ch211-288d18.1 | 0.130 | pnp4b | -0.084 |
| uncx | 0.127 | g6pca.2 | -0.084 |
| si:ch73-368j24.14 | 0.119 | cx32.3 | -0.084 |
| fgfr2 | 0.119 | ahcy | -0.083 |
| celf2 | 0.117 | apoc2 | -0.083 |
| si:ch211-268b14.2 | 0.116 | rdh1 | -0.083 |
| CU634008.1 | 0.116 | agxtb | -0.082 |
| foxp4 | 0.116 | phyhd1 | -0.082 |
| agrn | 0.116 | grhprb | -0.082 |
| myofl | 0.114 | apoa1b | -0.081 |
| elavl3 | 0.114 | eno3 | -0.080 |
| rbfox1 | 0.114 | bhmt | -0.080 |
| fam131bb | 0.113 | suclg2 | -0.080 |
| LOC101886926 | 0.113 | agxta | -0.080 |
| sparc | 0.112 | slco1d1 | -0.078 |
| marcksl1a | 0.112 | ces2 | -0.078 |
| spega | 0.112 | nipsnap3a | -0.078 |
| col18a1a | 0.111 | fabp10a | -0.078 |
| XLOC-002533 | 0.111 | tdo2a | -0.077 |
| ahnak | 0.110 | aqp12 | -0.077 |
| ptprn2 | 0.110 | acmsd | -0.076 |