MIA SH3 domain containing
ZFIN 
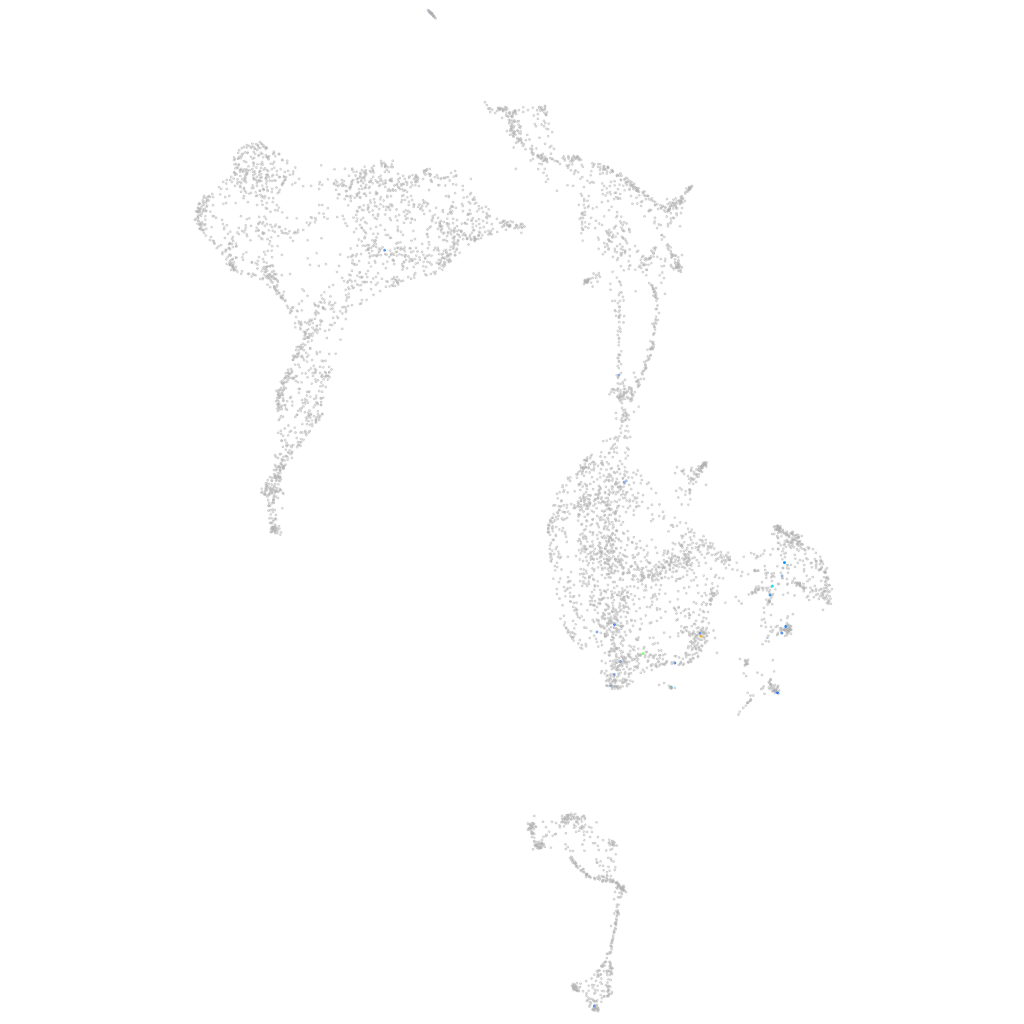
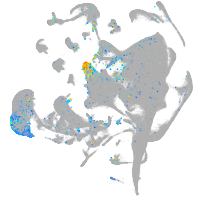
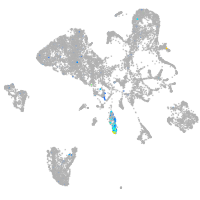
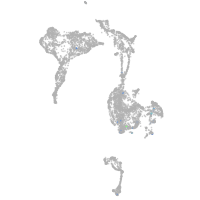
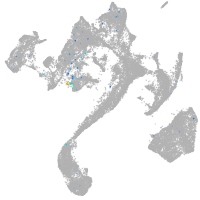
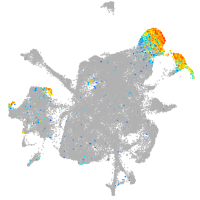
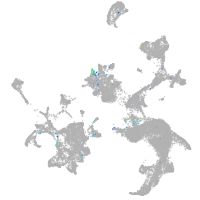
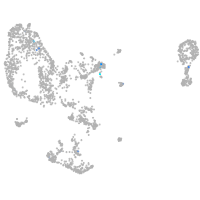
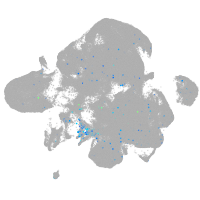
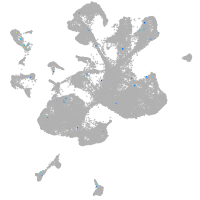
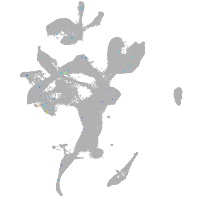
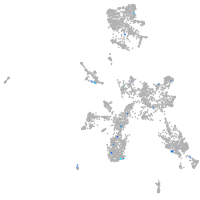
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| itih1 | 0.660 | sec61b | -0.054 |
| pcolceb | 0.547 | NC-002333.4 | -0.045 |
| matn1 | 0.476 | asph | -0.042 |
| pcdh2g8 | 0.417 | ptges3b | -0.040 |
| cthrc1b | 0.399 | hspb1 | -0.040 |
| CR759845.3 | 0.340 | tram1 | -0.040 |
| kcng4b | 0.340 | dnajc1 | -0.040 |
| abcc6b.1 | 0.340 | ddx39ab | -0.039 |
| LOC100535907 | 0.340 | anp32a | -0.039 |
| si:ch211-176g13.8 | 0.333 | anp32b | -0.038 |
| tnxba | 0.305 | hmga1a | -0.037 |
| otos | 0.301 | calr3b | -0.037 |
| hspb2 | 0.278 | hnrnpa1a | -0.037 |
| col9a1a | 0.273 | etf1b | -0.037 |
| LOC100330861 | 0.262 | eif4e1c | -0.036 |
| CYTL1 | 0.240 | npm1a | -0.036 |
| fgfbp2b | 0.227 | kdelr2a | -0.035 |
| zgc:172133 | 0.226 | odc1 | -0.035 |
| prlh2 | 0.226 | hnrnpa1b | -0.035 |
| gdf10a | 0.225 | ppig | -0.035 |
| CR854945.1 | 0.224 | chaf1a | -0.035 |
| tph2 | 0.219 | sart1 | -0.035 |
| opn1sw2 | 0.217 | nop58 | -0.034 |
| VIT | 0.215 | ssr1 | -0.034 |
| LOC101884915 | 0.213 | zgc:110216 | -0.034 |
| ntsr1 | 0.213 | ncl | -0.034 |
| XLOC-011687 | 0.213 | dkc1 | -0.034 |
| cabp4 | 0.213 | hnrnpaba | -0.034 |
| loxl4 | 0.203 | top1l | -0.033 |
| mhc1lfa | 0.197 | akap12b | -0.033 |
| dlgap2b | 0.195 | snrnp70 | -0.033 |
| CABZ01054023.1 | 0.195 | rrbp1a | -0.033 |
| hmx3b | 0.190 | fbl | -0.033 |
| myrf | 0.189 | snrpa1 | -0.033 |
| XLOC-020232 | 0.189 | syncrip | -0.033 |