milk fat globule-EGF factor 8 protein a
ZFIN 
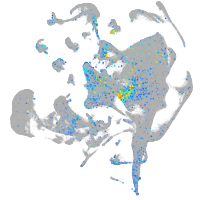
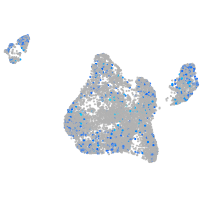
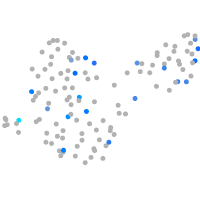
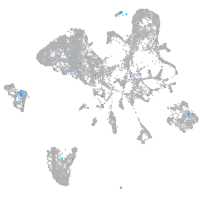
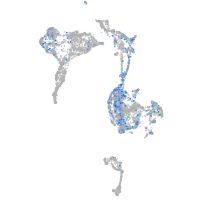
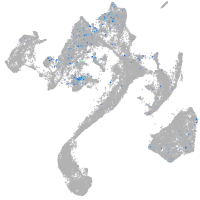
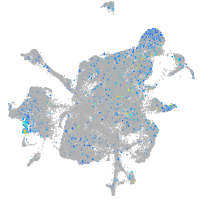
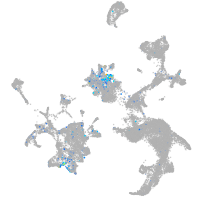
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| entpd3 | 0.304 | NC-002333.4 | -0.148 |
| shha | 0.285 | tmsb | -0.137 |
| BX323559.3 | 0.272 | elavl3 | -0.137 |
| gpc1b | 0.272 | stmn1b | -0.108 |
| zgc:163121 | 0.265 | zgc:55461 | -0.104 |
| slc13a5b | 0.261 | sp8a | -0.101 |
| cldni | 0.260 | pvalb5 | -0.101 |
| sostdc1a | 0.259 | ebf3a | -0.100 |
| LOC100536671 | 0.258 | spata4 | -0.100 |
| dpep1 | 0.255 | si:ch211-153b23.5 | -0.099 |
| si:cabz01007794.1 | 0.254 | hsbp1a | -0.098 |
| ccnd2a | 0.253 | atf5a | -0.098 |
| adgrf3b | 0.252 | uncx | -0.097 |
| LOC100003133 | 0.248 | tuba1a | -0.097 |
| foxq1a | 0.248 | tspan2a | -0.096 |
| apoeb | 0.247 | bcl11ba | -0.095 |
| XLOC-039931 | 0.247 | tmeff1b | -0.094 |
| cd44a | 0.247 | LOC100537342 | -0.093 |
| si:rp71-77l1.1 | 0.246 | capslb | -0.092 |
| pkd1l3 | 0.243 | rtn1a | -0.092 |
| crip1 | 0.243 | insm1a | -0.091 |
| si:ch211-149b19.4 | 0.238 | zgc:65894 | -0.091 |
| atp2a3 | 0.237 | meig1 | -0.090 |
| si:busm1-71b9.3 | 0.237 | vdac3 | -0.088 |
| igfbp7 | 0.236 | marcksb | -0.086 |
| gng13a | 0.235 | sp8b | -0.085 |
| trpm5 | 0.234 | dlx5a | -0.084 |
| CR318588.3 | 0.234 | olfm1b | -0.084 |
| LO018550.2 | 0.234 | vsig8a | -0.083 |
| kcnk17 | 0.233 | nova2 | -0.083 |
| ceacam1 | 0.232 | si:dkeyp-75h12.5 | -0.083 |
| sost | 0.232 | XLOC-024343 | -0.082 |
| si:ch73-199e17.1 | 0.232 | nkain1 | -0.082 |
| capgb | 0.232 | apex1 | -0.081 |
| klf3 | 0.231 | krt18b | -0.081 |