mesenchyme homeobox 1
ZFIN 
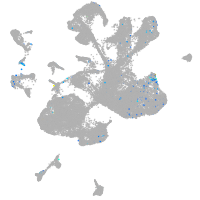
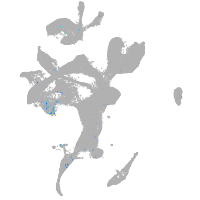
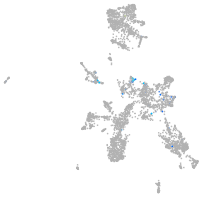
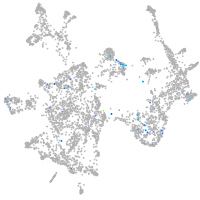
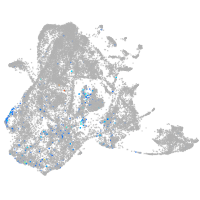
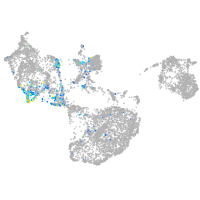
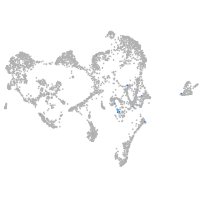
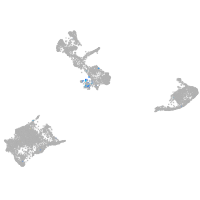
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| col6a2 | 0.279 | syngr1a | -0.051 |
| thbs4b | 0.274 | smim29 | -0.050 |
| LOC108179343 | 0.268 | impdh1b | -0.040 |
| mmp14a | 0.255 | agtrap | -0.039 |
| crygm2d16 | 0.250 | slc2a15a | -0.037 |
| twist1a | 0.248 | gmps | -0.036 |
| ms4a17a.9 | 0.240 | rab32a | -0.036 |
| rbp7b | 0.239 | LOC103910009 | -0.035 |
| tril | 0.238 | gpr143 | -0.035 |
| col6a1 | 0.238 | tpd52 | -0.035 |
| pdgfrl | 0.237 | si:dkey-21a6.5 | -0.034 |
| vwde | 0.235 | atp11a | -0.033 |
| c1qtnf2 | 0.233 | gstp1 | -0.033 |
| LOC110438394 | 0.231 | LOC103909099 | -0.033 |
| col5a1 | 0.231 | rab38 | -0.033 |
| col5a2a | 0.226 | slc45a2 | -0.033 |
| fap | 0.225 | rnaseka | -0.032 |
| clcf1 | 0.221 | tspan36 | -0.032 |
| krt18a.1 | 0.220 | bloc1s6 | -0.032 |
| mfap2 | 0.219 | emp3b | -0.032 |
| si:ch1073-459j12.1 | 0.213 | bace2 | -0.032 |
| lrrc17 | 0.212 | trpm1b | -0.032 |
| dcn | 0.210 | ivns1abpa | -0.031 |
| serpinf1 | 0.210 | nme4 | -0.031 |
| si:ch73-24k9.2 | 0.208 | aldh16a1 | -0.031 |
| col1a2 | 0.205 | rabl6b | -0.031 |
| fam198a | 0.203 | mtbl | -0.030 |
| fmoda | 0.202 | rhag | -0.030 |
| scara3 | 0.202 | kcnj13 | -0.030 |
| si:dkey-12l12.1 | 0.198 | cmtm7 | -0.029 |
| si:ch211-215p11.1 | 0.198 | prps1a | -0.029 |
| col1a1b | 0.197 | gm2a | -0.029 |
| dnase1l1l | 0.196 | uraha | -0.029 |
| mespba | 0.196 | arhgef33 | -0.029 |
| zgc:173915 | 0.196 | paics | -0.029 |