multiple EGF-like-domains 8
ZFIN 
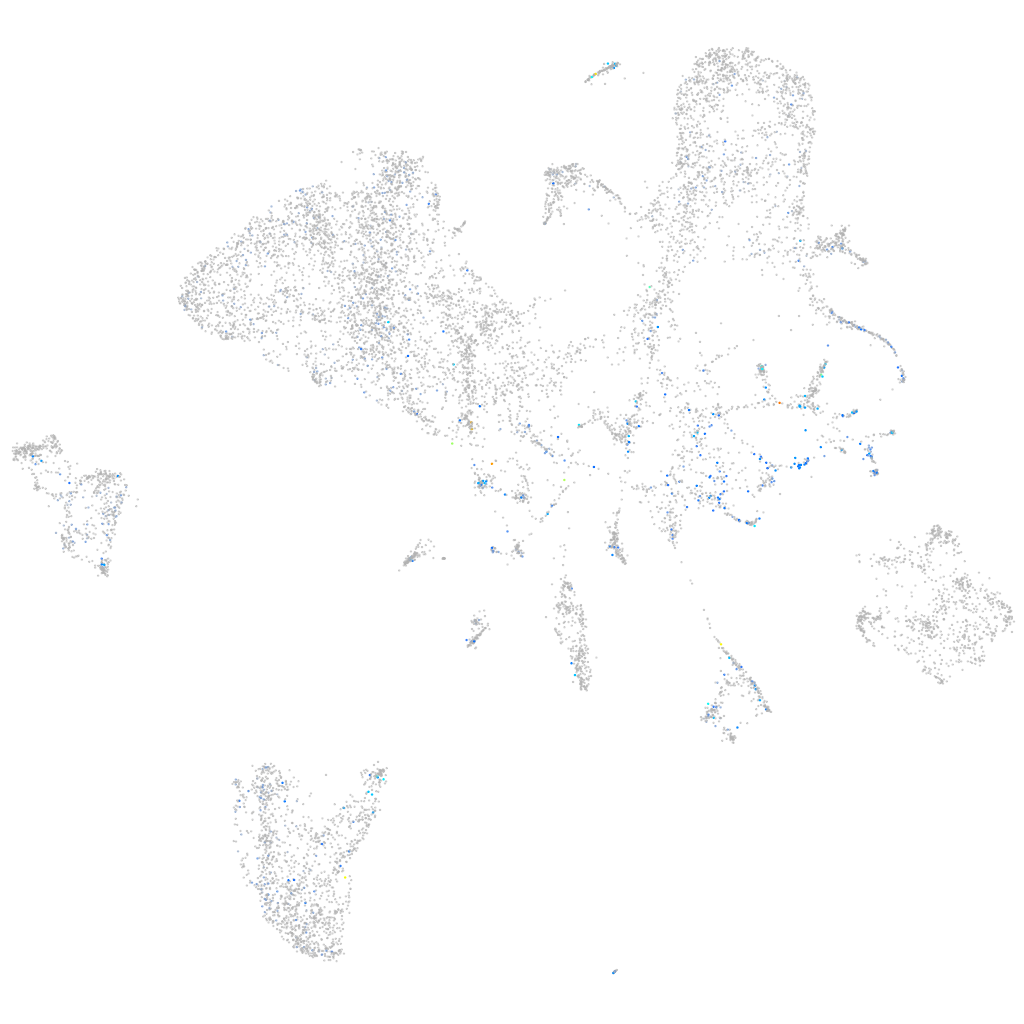
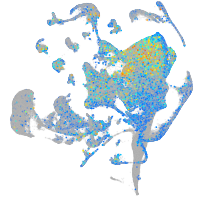
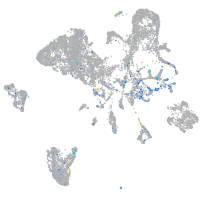
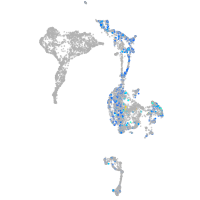
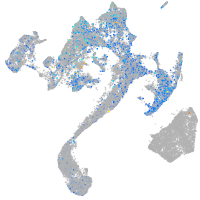
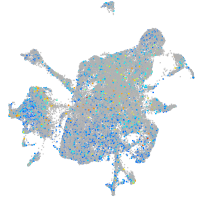
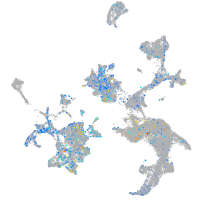
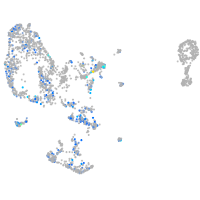
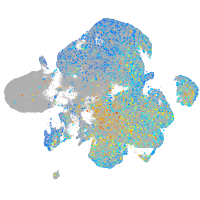
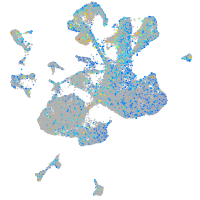
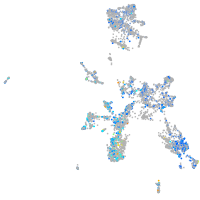
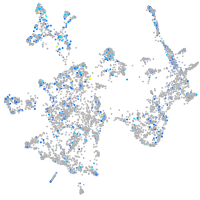
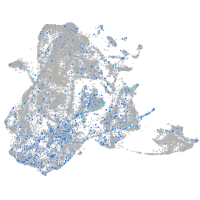
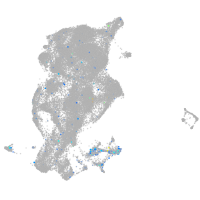
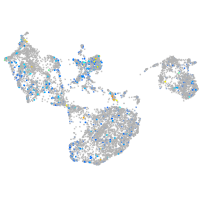
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| FO818719.1 | 0.185 | aldob | -0.097 |
| CR392036.1 | 0.156 | gapdh | -0.096 |
| BX936415.1 | 0.155 | ahcy | -0.089 |
| XLOC-006820 | 0.150 | eno3 | -0.085 |
| ptchd4 | 0.149 | apoc2 | -0.085 |
| syt1a | 0.148 | gamt | -0.084 |
| nrxn1a | 0.145 | apoa4b.1 | -0.082 |
| NPFFR2 | 0.144 | afp4 | -0.078 |
| lysmd2 | 0.138 | fbp1b | -0.078 |
| si:ch73-233f7.6 | 0.136 | gpx4a | -0.078 |
| scg3 | 0.135 | gstt1a | -0.077 |
| vamp2 | 0.134 | gatm | -0.076 |
| LOC103910736 | 0.134 | sult2st2 | -0.075 |
| tox | 0.132 | scp2a | -0.074 |
| CR383676.1 | 0.132 | apoa1b | -0.073 |
| gpr88 | 0.132 | ugt1a7 | -0.073 |
| mir29b-1 | 0.130 | suclg2 | -0.069 |
| lingo2a | 0.129 | gcshb | -0.069 |
| chga | 0.128 | glud1b | -0.069 |
| ssx2ipb | 0.128 | rdh1 | -0.069 |
| camk1ga | 0.128 | fabp2 | -0.068 |
| trpc6b | 0.125 | apobb.1 | -0.068 |
| appb | 0.124 | apoc1 | -0.067 |
| neurod1 | 0.124 | sod1 | -0.066 |
| zgc:114175 | 0.124 | suclg1 | -0.065 |
| BX005419.1 | 0.123 | gstr | -0.065 |
| CU915256.2 | 0.123 | rbp2a | -0.064 |
| cacng2a | 0.121 | msrb2 | -0.064 |
| sdcbp | 0.121 | pla2g12b | -0.063 |
| gdi1 | 0.121 | mat1a | -0.063 |
| ppp1r14ba | 0.121 | agxta | -0.063 |
| si:ch1073-429i10.3.1 | 0.121 | cx32.3 | -0.063 |
| insm1a | 0.120 | ugt5b4 | -0.062 |
| gpc1a | 0.120 | cyp2n13 | -0.062 |
| cxxc5a | 0.119 | sult1st6 | -0.062 |