myocyte enhancer factor 2d
ZFIN 
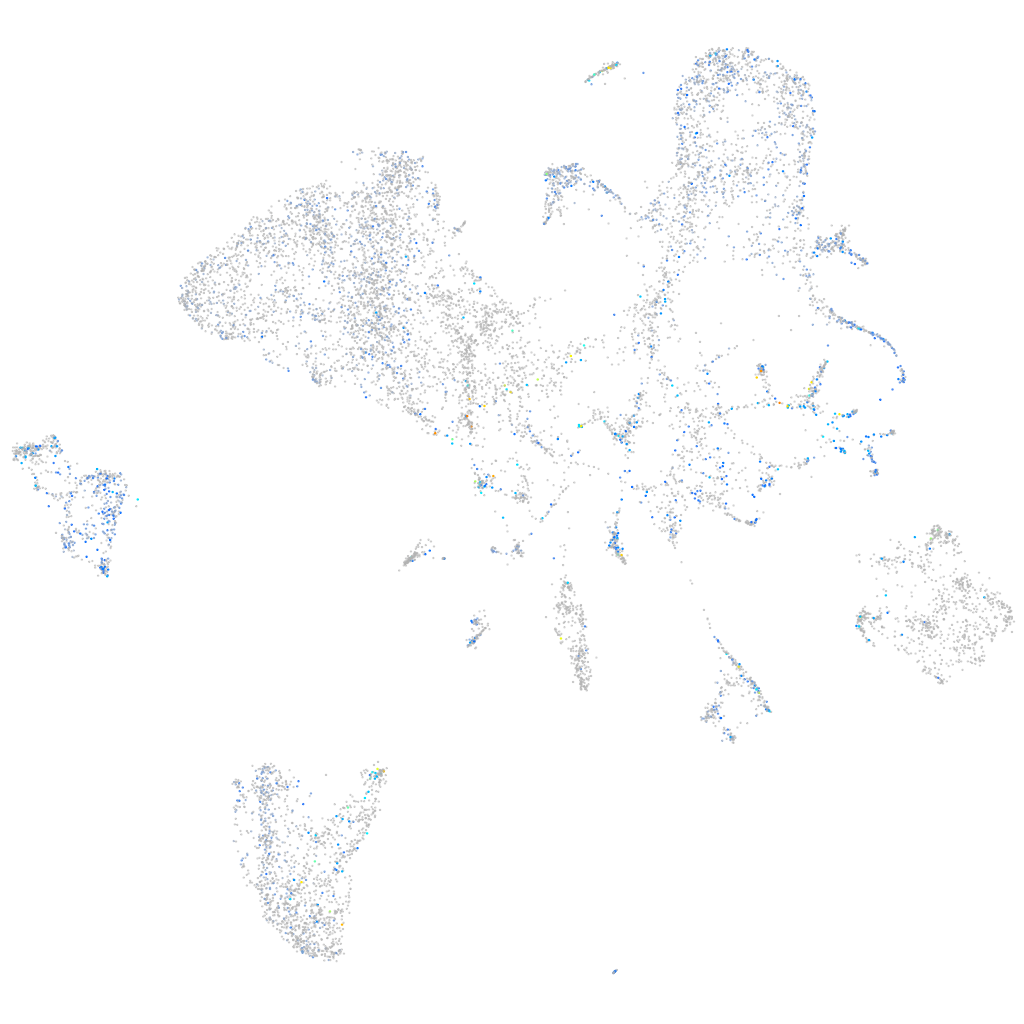
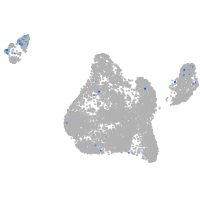
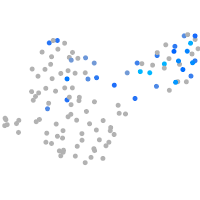
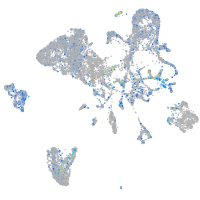
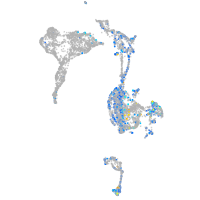
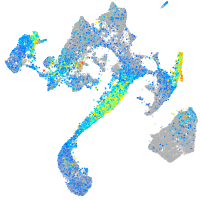
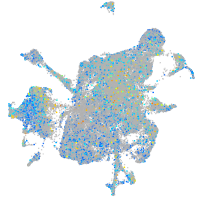
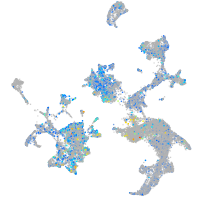
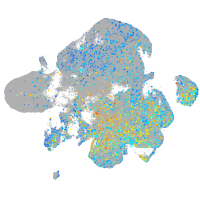
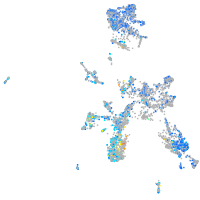
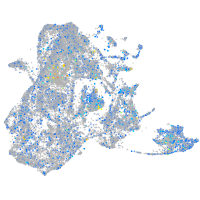
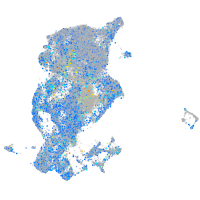
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| CR383676.1 | 0.176 | apoc1 | -0.127 |
| btg1 | 0.154 | bhmt | -0.120 |
| mcl1a | 0.143 | gamt | -0.119 |
| cldnh | 0.142 | apoa1b | -0.109 |
| si:ch1073-429i10.3.1 | 0.141 | si:dkey-16p21.8 | -0.106 |
| ier2b | 0.140 | apoa2 | -0.105 |
| gapdhs | 0.139 | gatm | -0.105 |
| calm1a | 0.139 | serpina1 | -0.103 |
| jun | 0.137 | apom | -0.103 |
| syt1a | 0.137 | ahcy | -0.101 |
| rnasekb | 0.136 | grhprb | -0.101 |
| epcam | 0.135 | tfa | -0.100 |
| icn | 0.133 | hao1 | -0.100 |
| si:dkey-153k10.9 | 0.133 | gapdh | -0.100 |
| vat1 | 0.133 | rbp2b | -0.098 |
| calm1b | 0.131 | zgc:123103 | -0.098 |
| scg3 | 0.130 | serpina1l | -0.098 |
| tmed7 | 0.129 | fetub | -0.098 |
| ccni | 0.129 | agxtb | -0.098 |
| pkma | 0.128 | fgg | -0.098 |
| tmem59 | 0.128 | mat1a | -0.097 |
| gnb1a | 0.127 | afp4 | -0.097 |
| ildr1a | 0.126 | ambp | -0.096 |
| sat1a.2 | 0.125 | ttc36 | -0.096 |
| atf4b | 0.125 | ftcd | -0.095 |
| cap1 | 0.125 | aqp12 | -0.095 |
| ptbp3 | 0.124 | fgb | -0.094 |
| neurod1 | 0.123 | uox | -0.094 |
| si:ch211-196l7.4 | 0.123 | ttr | -0.093 |
| pcsk1 | 0.123 | fabp10a | -0.093 |
| kif5aa | 0.123 | ces2 | -0.093 |
| kcnj11 | 0.122 | agxta | -0.093 |
| kdm6bb | 0.122 | cpn1 | -0.093 |
| h1fx | 0.122 | LOC110437731 | -0.092 |
| ier5 | 0.121 | fga | -0.092 |