mucolipin 2
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm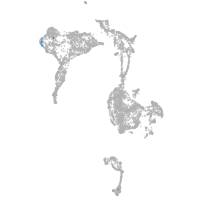 |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature 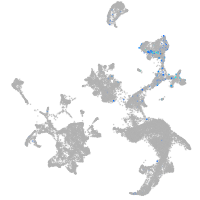 |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line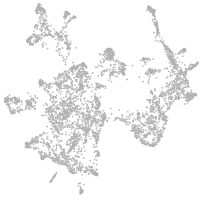 |
epidermis |
periderm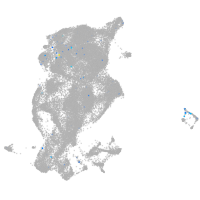 |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ccdc175 | 0.526 | NC-002333.4 | -0.034 |
| LOC100536937 | 0.526 | vps26c | -0.033 |
| znf1166 | 0.526 | ndufs3 | -0.032 |
| CU550719.2 | 0.526 | ndufv2 | -0.032 |
| si:dkey-222b8.4 | 0.504 | atf4a | -0.031 |
| dnaaf3l | 0.500 | mrpl35 | -0.030 |
| lrguk | 0.487 | pdha1a | -0.030 |
| CU633857.1 | 0.465 | phb2b | -0.030 |
| wdr27 | 0.448 | calm1a | -0.030 |
| si:ch1073-181h11.2 | 0.438 | ndufa9a | -0.030 |
| si:dkeyp-86c4.1 | 0.431 | ndufb9 | -0.030 |
| rsph3 | 0.424 | rgs4 | -0.029 |
| cfap300 | 0.422 | atp6v1d | -0.029 |
| cfap100 | 0.421 | ndufv3 | -0.029 |
| c18h15orf65 | 0.398 | atp6v1f | -0.029 |
| got1l1 | 0.389 | bckdhb | -0.028 |
| drc1 | 0.387 | ndufb10 | -0.028 |
| spata18 | 0.381 | afap1l2 | -0.028 |
| CR855277.2 | 0.370 | ufc1 | -0.028 |
| mustn1a | 0.370 | coq9 | -0.028 |
| lrrc51 | 0.368 | sat1a.1 | -0.028 |
| ccl32b.3 | 0.359 | btg1 | -0.027 |
| dnali1 | 0.339 | si:dkey-88p24.11 | -0.027 |
| tbata | 0.330 | degs1 | -0.027 |
| znf1016 | 0.328 | mrpl19 | -0.027 |
| ccdc146 | 0.326 | cib1 | -0.027 |
| ccdc13 | 0.321 | exoc1 | -0.027 |
| si:dkeyp-69c1.9 | 0.318 | ndufb5 | -0.027 |
| nme5 | 0.307 | dido1 | -0.027 |
| katnal2 | 0.297 | pdxdc1 | -0.027 |
| tekt1 | 0.294 | rnaseka | -0.027 |
| foxj1b | 0.290 | fscn1a | -0.027 |
| dnaaf4 | 0.289 | ppp3r1b | -0.027 |
| si:dkey-163f12.6 | 0.287 | rraga | -0.027 |
| cd37 | 0.282 | ndufa4 | -0.026 |