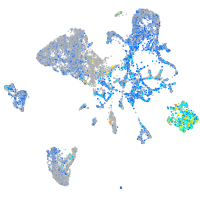
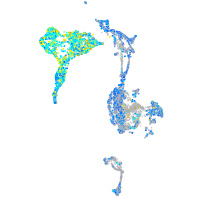
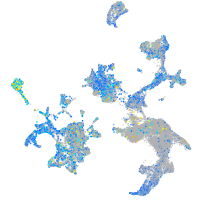
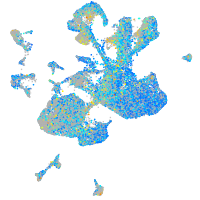
matrin 3-like 1.1
ZFIN 
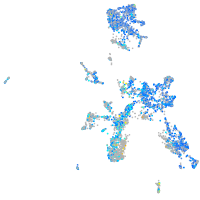
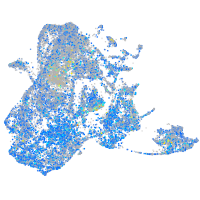
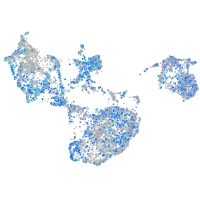
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| seta | 0.611 | ptmaa | -0.513 |
| chaf1a | 0.590 | ppiab | -0.487 |
| cx43.4 | 0.565 | ccni | -0.471 |
| hnrnpub | 0.561 | pvalb1 | -0.462 |
| zc3h13 | 0.559 | mdh2 | -0.462 |
| syncrip | 0.557 | h3f3a | -0.453 |
| ddx4 | 0.552 | rpl37 | -0.452 |
| sf3b4 | 0.546 | rnaset2 | -0.446 |
| kri1 | 0.534 | pvalb2 | -0.443 |
| khdrbs1b | 0.524 | ndrg3a | -0.439 |
| pes | 0.523 | rnasekb | -0.436 |
| ppig | 0.511 | mylpfa | -0.433 |
| nkap | 0.511 | anxa4 | -0.429 |
| ube2nb | 0.509 | rps27.1 | -0.428 |
| zgc:56699 | 0.509 | si:ch211-191j22.3 | -0.426 |
| nasp | 0.506 | b2ml | -0.424 |
| hnrnpabb | 0.504 | litaf | -0.424 |
| ube2na | 0.498 | rps23 | -0.424 |
| sde2 | 0.496 | icn | -0.423 |
| stmn1a | 0.495 | scgn | -0.422 |
| h1m | 0.486 | dap1b | -0.421 |
| NC-002333.4 | 0.483 | rpl9 | -0.420 |
| ISCU | 0.477 | cox8a | -0.420 |
| rnf20 | 0.472 | rps15 | -0.415 |
| hnrnph1l | 0.466 | vdac2 | -0.413 |
| dnajc1 | 0.465 | rtn1a | -0.409 |
| ncaph | 0.461 | rps17 | -0.408 |
| pdap1a | 0.458 | rps10 | -0.407 |
| mki67 | 0.457 | cyt1 | -0.404 |
| baz1b | 0.457 | atp5f1b | -0.404 |
| nop2 | 0.456 | ldhba | -0.399 |
| anp32b | 0.454 | gapdhs | -0.398 |
| mphosph10 | 0.454 | calm1a | -0.396 |
| AL935174.5 | 0.449 | acat1 | -0.396 |
| hmgb2a | 0.449 | rplp0 | -0.393 |