microtubule associated serine/threonine kinase 1b
ZFIN 
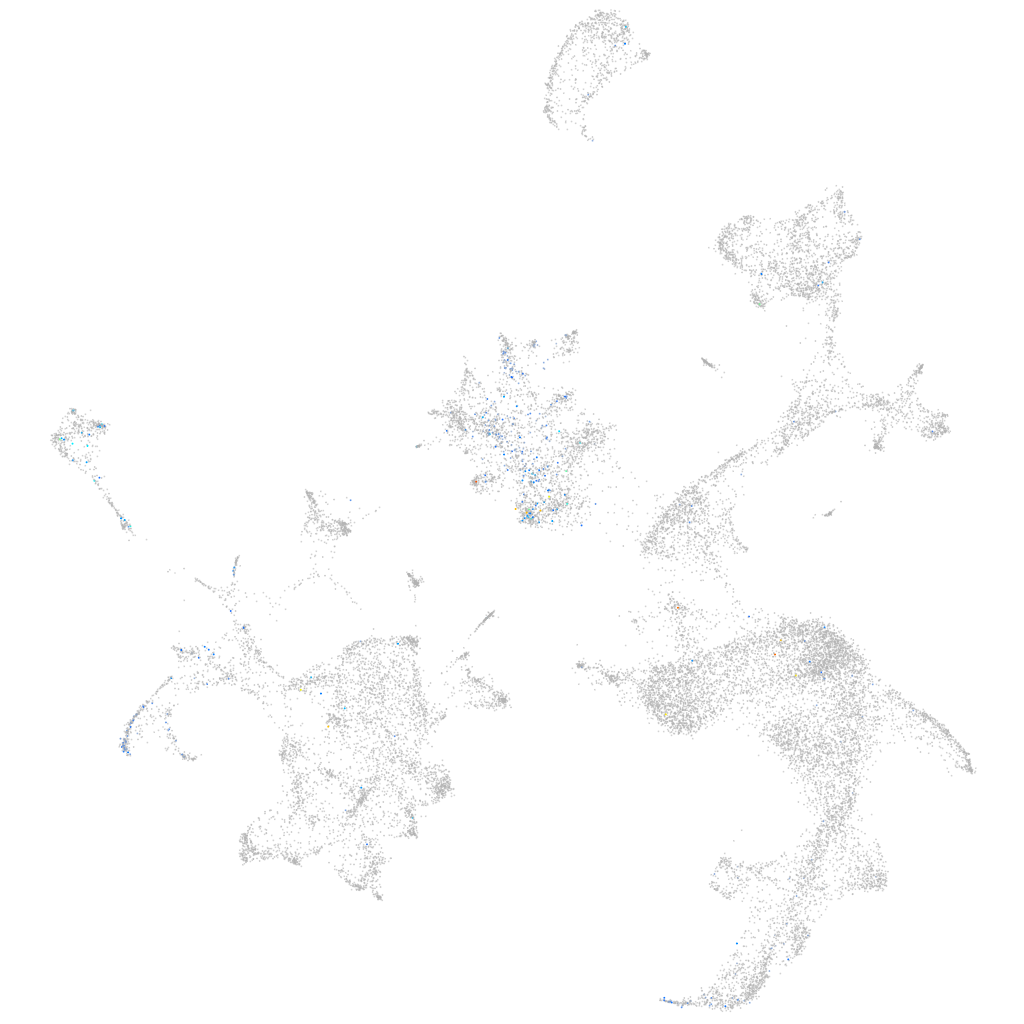
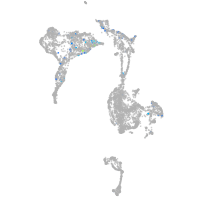
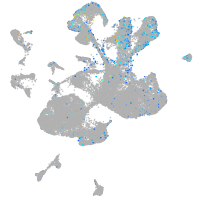
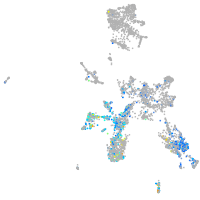
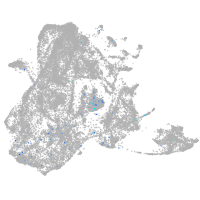
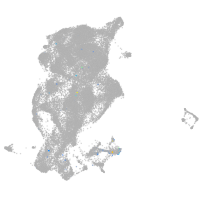
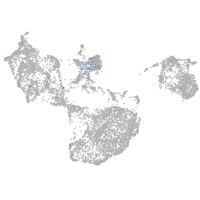
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| erc1b | 0.165 | srgn | -0.036 |
| cldn23b | 0.148 | pvalb1 | -0.031 |
| LOC103911284 | 0.141 | b3gnt3.4 | -0.027 |
| stmn1b | 0.134 | ecscr | -0.026 |
| rpp25b | 0.134 | si:ch73-334d15.2 | -0.026 |
| sncb | 0.131 | kdrl | -0.025 |
| kcng4b | 0.127 | coro1a | -0.025 |
| gpm6aa | 0.126 | hlx1 | -0.024 |
| tuba1c | 0.123 | rac2 | -0.024 |
| nxph2a | 0.121 | arpc1b | -0.024 |
| gng3 | 0.120 | capgb | -0.024 |
| dnajc5aa | 0.118 | ponzr1 | -0.024 |
| elavl3 | 0.117 | rps17 | -0.024 |
| atp6v0cb | 0.117 | tagln2 | -0.024 |
| elavl4 | 0.116 | gmfg | -0.024 |
| tuba1a | 0.114 | laptm5 | -0.024 |
| vamp2 | 0.113 | mcl1a | -0.024 |
| dnm1a | 0.112 | cdh5 | -0.023 |
| syt11b | 0.111 | pvalb2 | -0.023 |
| rtn1a | 0.111 | tmod4 | -0.023 |
| pitpnab | 0.111 | XLOC-039757 | -0.023 |
| atpv0e2 | 0.111 | pecam1 | -0.023 |
| atp1b2a | 0.110 | gsto2 | -0.023 |
| ywhag2 | 0.109 | eno3 | -0.023 |
| sez6l2 | 0.109 | si:ch211-156j16.1 | -0.022 |
| syn2a | 0.109 | thsd1 | -0.022 |
| si:ch211-214j24.9 | 0.109 | si:ch211-248e11.2 | -0.022 |
| kif1ab | 0.108 | plk2b | -0.022 |
| ppfia3 | 0.108 | cldn5b | -0.022 |
| cspg5a | 0.108 | myct1a | -0.022 |
| cdk5r1b | 0.108 | sox18 | -0.022 |
| glrbb | 0.107 | arhgdig | -0.022 |
| sv2a | 0.107 | crip1 | -0.022 |
| myo16 | 0.106 | dub | -0.022 |
| si:ch211-137a8.4 | 0.106 | aqp1a.1 | -0.021 |