mitogen-activated protein kinase 15
ZFIN 
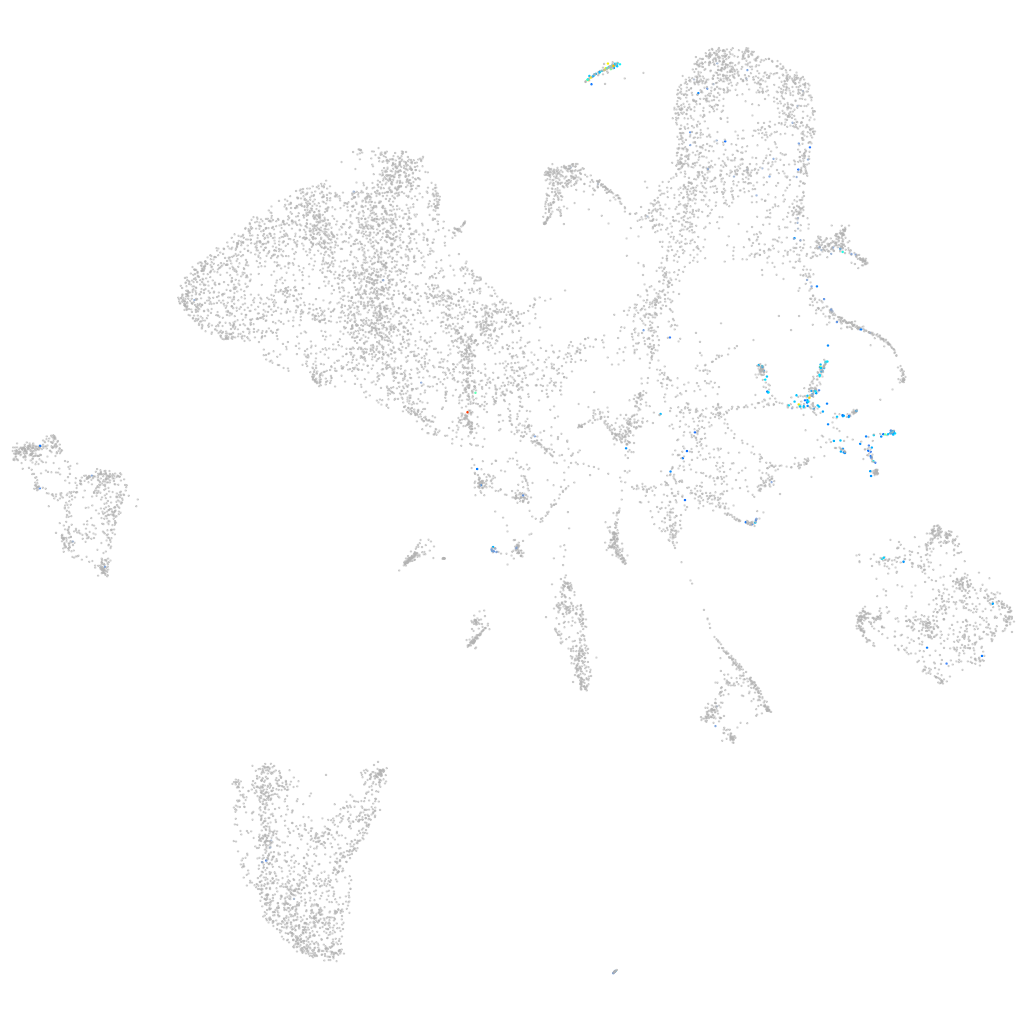
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| scg3 | 0.463 | ahcy | -0.132 |
| zgc:101731 | 0.406 | gapdh | -0.127 |
| neurod1 | 0.382 | gamt | -0.123 |
| c2cd4a | 0.379 | fabp3 | -0.115 |
| fev | 0.379 | gatm | -0.112 |
| pcsk1nl | 0.349 | bhmt | -0.106 |
| insm1b | 0.344 | aldob | -0.105 |
| gpc1a | 0.340 | mat1a | -0.102 |
| egr4 | 0.332 | apoa4b.1 | -0.101 |
| mir7a-1 | 0.332 | aldh6a1 | -0.100 |
| scgn | 0.320 | fbp1b | -0.100 |
| syt1a | 0.318 | apoc2 | -0.099 |
| slc7a14a | 0.311 | hdlbpa | -0.099 |
| adcyap1a | 0.311 | eif4ebp1 | -0.098 |
| hepacam2 | 0.309 | cx32.3 | -0.098 |
| si:dkey-153k10.9 | 0.306 | aldh7a1 | -0.098 |
| mir375-2 | 0.305 | afp4 | -0.097 |
| pcsk1 | 0.303 | apoa1b | -0.096 |
| pax6b | 0.302 | si:dkey-16p21.8 | -0.096 |
| pcsk2 | 0.301 | gstt1a | -0.095 |
| vat1 | 0.301 | nupr1b | -0.094 |
| scg2a | 0.288 | adka | -0.092 |
| tspan7b | 0.288 | aqp12 | -0.091 |
| gfra3 | 0.283 | gstr | -0.090 |
| nmbb | 0.283 | shmt1 | -0.090 |
| gabra2a | 0.283 | agxtb | -0.089 |
| kcnj11 | 0.282 | apoc1 | -0.089 |
| dll4 | 0.276 | scp2a | -0.089 |
| penka | 0.276 | agxta | -0.087 |
| lysmd2 | 0.275 | tfa | -0.087 |
| lmx1ba | 0.274 | wu:fj16a03 | -0.086 |
| rprmb | 0.272 | mgst1.2 | -0.086 |
| rasd4 | 0.266 | hspd1 | -0.086 |
| slc35g2a | 0.265 | cdo1 | -0.085 |
| alpk3b | 0.265 | apobb.1 | -0.085 |