mitogen-activated protein kinase 12a
ZFIN 
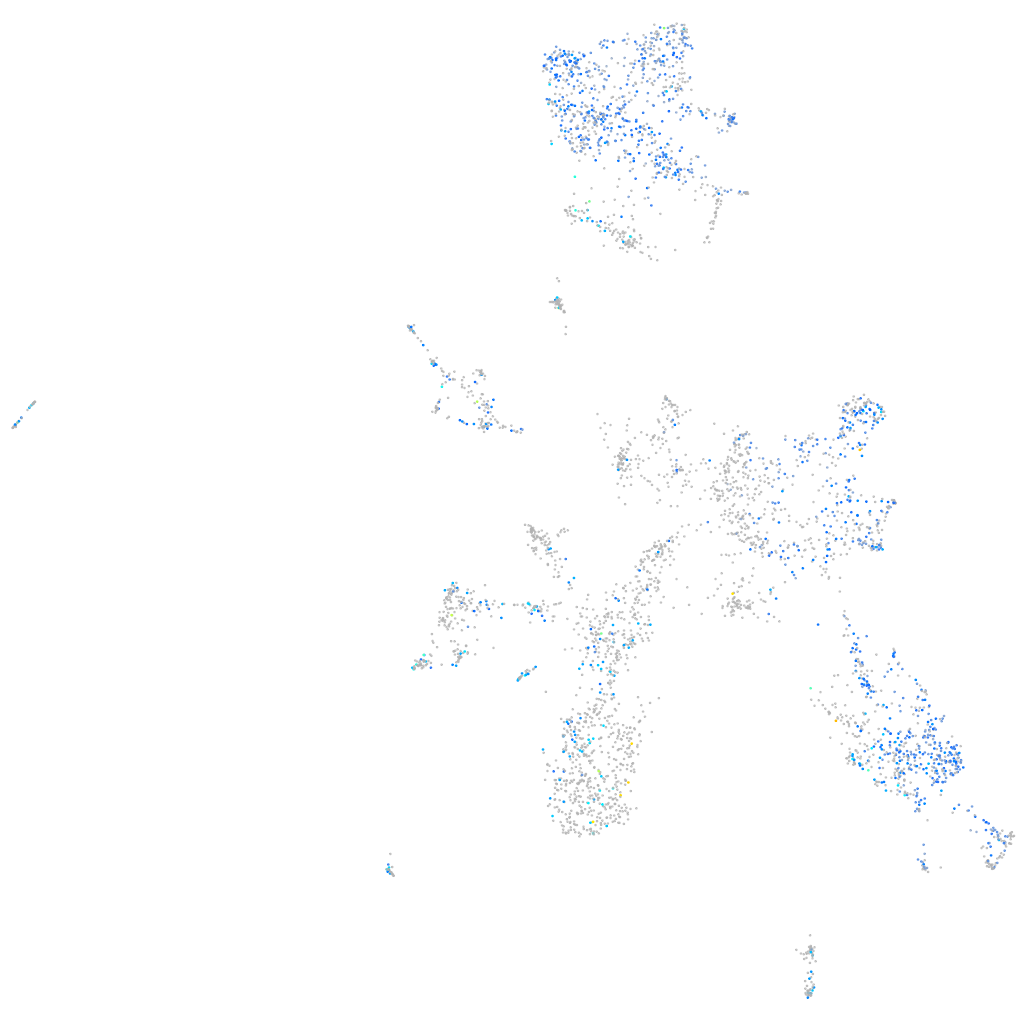
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cfl1l | 0.239 | elavl3 | -0.153 |
| zgc:158343 | 0.232 | si:dkey-56m19.5 | -0.138 |
| tmsb1 | 0.226 | CU467822.1 | -0.136 |
| krt8 | 0.225 | ebf2 | -0.127 |
| aqp3a | 0.222 | neurod4 | -0.123 |
| pfn1 | 0.222 | sox11a | -0.113 |
| cd9b | 0.215 | ebf3a | -0.113 |
| lgals3b | 0.213 | hbbe1.3 | -0.109 |
| anxa4 | 0.207 | tmeff1b | -0.108 |
| si:dkey-16l2.20 | 0.203 | sox11b | -0.106 |
| s100a10b | 0.202 | cxcr4b | -0.105 |
| krt18a.1 | 0.202 | neurod1 | -0.103 |
| GCA | 0.201 | myt1a | -0.100 |
| cotl1 | 0.200 | lhx2a | -0.098 |
| ahnak | 0.200 | tp53inp2 | -0.096 |
| fgfbp1b | 0.199 | tuba1a | -0.095 |
| sdc4 | 0.198 | nova2 | -0.094 |
| fbp1a | 0.197 | stmn1b | -0.094 |
| anxa11b | 0.196 | uncx | -0.092 |
| gpa33a | 0.195 | hmgb1b | -0.090 |
| phlda2 | 0.194 | insm1b | -0.090 |
| her9 | 0.194 | her4.2 | -0.089 |
| tagln2 | 0.194 | epb41a | -0.087 |
| s100v1 | 0.193 | si:ch211-193l2.6 | -0.087 |
| krt97 | 0.191 | NC-002333.4 | -0.087 |
| ier2b | 0.190 | LOC100537384 | -0.086 |
| zgc:101744 | 0.190 | ptmab | -0.084 |
| krt4 | 0.190 | lfng | -0.084 |
| cnn2 | 0.189 | dla | -0.081 |
| capns1a | 0.187 | hbae3 | -0.080 |
| spint1a | 0.187 | LOC100537342 | -0.079 |
| sparc | 0.186 | ebf1a | -0.079 |
| net1 | 0.186 | nono | -0.077 |
| cd63 | 0.186 | insm1a | -0.077 |
| pycard | 0.185 | rcor2 | -0.077 |